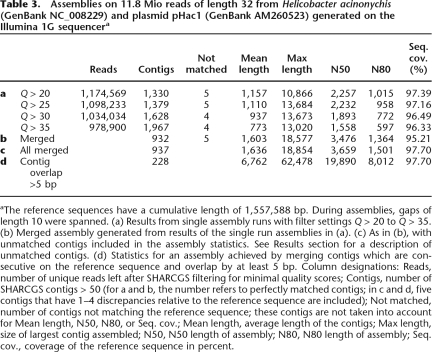
Table 3.
Assemblies on 11.8 Mio reads of length 32 from Helicobacter acinonychis (GenBank NC_008229) and plasmid pHac1 (GenBank AM260523) generated on the Illumina 1G sequencera
aThe reference sequences have a cumulative length of 1,557,588 bp. During assemblies, gaps of length 10 were spanned. (a) Results from single assembly runs with filter settings Q > 20 to Q > 35. (b) Merged assembly generated from results of the single run assemblies in (a). (c) As in (b), with unmatched contigs included in the assembly statistics. See Results section for a description of unmatched contigs. (d) Statistics for an assembly achieved by merging contigs which are consecutive on the reference sequence and overlap by at least 5 bp. Column designations: Reads, number of unique reads left after SHARCGS filtering for minimal quality scores; Contigs, number of SHARCGS contigs > 50 (for a and b, the number refers to perfectly matched contigs; in c and d, five contigs that have 1–4 discrepancies relative to the reference sequence are included); Not matched, number of contigs not matching the reference sequence; these contigs are not taken into account for Mean length, N50, N80, or Seq. cov.; Mean length, average length of the contigs; Max length, size of largest contig assembled; N50, N50 length of assembly; N80, N80 length of assembly; Seq. cov., coverage of the reference sequence in percent.