FIG. 1.
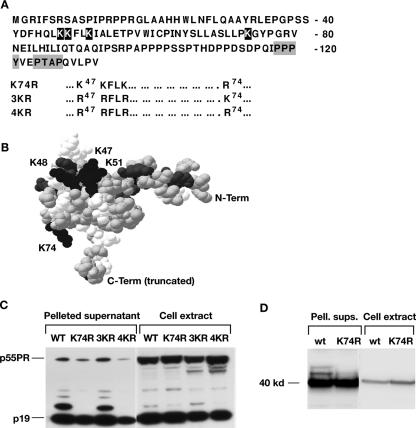
K74 is the site of HTLV-1 Gag ubiquitination. (A) Lysine residues in the HTLV-1 MA domain are indicated in white print, and the LD motifs, PPPY and PTAP, are boxed. The amino acid changes in the mutants K74R, 3KR and 4KR are shown underneath the MA sequence. (B) The structure of HTLV-1 MA was modeled by Swiss-Model based on the nuclear magnetic resonance structure of HTLV-2 MA (6). The lysine residues are labeled and shown in black; the other basic residues are shown in gray. The sequence was truncated at residue 100 for the purpose of modeling, as the C terminus is unstructured. (C) Immunoblot analysis of cell extracts and pelleted supernatants from 293T cultures transfected with pCMVHT-1ΔX, pCMVHT-1Δ-K74R, pCMVHT-1ΔX-3KR, and pCMVHT-1ΔX-4KR. Blots were probed with mouse monoclonal anti-HTLV-1 p19(MA) antibody. (D) Immunoblot analysis of cell extracts and pelleted supernatants (Pell. sups.) from 293T cells transfected with pCMV-MACA-RRE and pCMV-MACA-K74R-RRE. Blots were treated as described above.