Abstract
Tumor necrosis factor alpha (TNF-α) is believed to play a significant role in the pathogenesis of dengue virus (DV) infection, with elevated levels of TNF-α in the sera of DV-infected patients paralleling the severity of disease and TNF-α release being coincident with the peak of DV production from infected monocyte-derived macrophages (MDM) in vitro. Since macrophages are a primary cell target in vivo for DV infection, we investigated the potential antiviral role of TNF-α in regulating DV replication in MDM. While pretreatment of MDM with TNF-α had a minor inhibitory effect, addition of TNF-α to MDM with established DV infection had no effect on DV replication as measured by DV RNA levels or progeny virus production. Blocking endogenous TNF-α using short interfering RNA or inhibitory TNF-α antibodies also had no effect on infectious DV production or viral RNA synthesis. Together, these results demonstrate that DV replication in MDM is not affected by TNF-α. Additionally, normal cellular TNF-α signaling, measured by quantitation of TNF-α-induced stimulation of transcription from an NF-κB-responsive reporter plasmid or NF-κB protein nuclear translocation, was blocked in DV-infected MDM and Huh7 cells. Thus, DV replication in MDM is not affected by TNF-α, and infected cells do not respond normally to TNF-α stimulation. It is therefore unlikely that the increased production of TNF-α seen in DV infection directly effects DV clearance by reducing DV replication, and the ability of DV to alter TNF-α responsiveness highlights another example of viral subversion of cellular functions.
Dengue virus (DV) is a member of the family Flaviviridae, and DV infection in humans causes a wide range of diseases, from acute febrile illness (dengue fever) to life-threatening dengue hemorrhagic fever and dengue shock syndrome (57). Serious forms of the disease involving DV-induced shock and hemorrhage are thought to be immune mediated by vasoactive cytokines such as tumor necrosis factor alpha (TNF-α). DV has been shown to infect numerous cell lines in vitro including endothelial cells, B and T cells, and hepatocytes (5, 31, 34, 38). However, one important target cell type for DV infection in vivo is cells of the monocyte lineage, based on the recovery of live virus from this cell type from the circulation of DV-infected individuals and the identification of DV-infected cells in pathological specimens (23, 24, 55). In vitro infection of primary cells such as cultured primary macrophages (9, 24, 50), dendritic cells (26, 33, 39, 58), and B cells (35) probably more closely represents viral replication in the target cells involved in vivo.
Elevated levels of TNF-α are found in patients with DV infection, with even higher levels associated with more serious forms of the disease (19, 27, 30). Exposure of monocytes/macrophages to DV particles or proteins has been suggested to be responsible for the enhanced production of TNF-α in DV-infected patients (28). DV challenge of BALB/c mice (haplotype H-2d) with a mouse-adapted DV type 2 (DV2) strain results in clinical symptoms of DV disease, including hemorrhagic shock with a 100% mortality rate (3). In these DV-infected mice, levels of TNF-α are increased and treatment with anti-TNF-α antibody reduces the mortality rate to 40%. Recent studies using a new DV strain, D2S10, which mimics human DV disease including viremia and increased vascular permeability in mice, have confirmed that TNF-α is one of the key cytokines responsible for the severity of DV disease and lethality in mice (56). In vitro studies have shown that exposure of human endothelial cells to culture fluids from DV-infected monocytes or macrophages triggers endothelial cell activation (2, 7), which can be reversed by treatment with anti-TNF-α antibodies (2, 6). Together these data suggest that circulating TNF-α levels are high in DV infection and are involved in the pathogenesis of disease, potentially by direct effects on the endothelium; however, the effects of these high levels of TNF-α on DV replication are unknown.
TNF-α is a powerful proinflammatory cytokine with pleiotropic properties and can have antiviral effects on several viruses (reviewed in reference 25). For example, TNF-α is associated with viral clearance in hepatitis B virus (HBV)-infected transgenic mice by inhibiting HBV gene expression and accelerating the degradation of HBV mRNA (12, 18, 22), endogenously released TNF-α plays a part in cytomegalovirus (CMV) clearance (48), and TNF-α can suppress human immunodeficiency virus type 1 (HIV-1) replication in peripheral blood monocytes and alveolar macrophages (32) and can protect mice from infection with herpes simplex virus type 1 (52). Challenge of mouse embryonic fibroblasts with West Nile virus (WNV), another flavivirus, has shown that mouse embryonic fibroblasts from TNF-α-deficient mice are more susceptible to WNV infection than are cells from wild-type mice, suggesting a role of TNF-α in protection against initial WNV infection (11).
In relation to DV, however, possible antiviral effects of TNF-α on DV replication are poorly understood. TNF-α is released from monocyte-derived macrophages (MDM) after DV infection, and the peak of this release coincides with the peak of virus production in vitro (7, 16). Other cells of the immune system such as B and T cells that functionally interact with monocytes and macrophages during viral infection can also release TNF-α when exposed to DV (35-37). Thus, TNF-α released from cells of the immune system may contribute to the elevated circulating (endocrine) levels of TNF-α that may in turn induce viral pathogenesis (see above); in addition, TNF-α may act in an autocrine or paracrine manner to modulate viral replication in DV-infected cells.
In this study, we have used an in vitro primary cell culture system to examine whether increased levels of TNF-α could affect DV replication in macrophages. Our results showed that addition of exogenous TNF-α, or inhibition of endogenously produced TNF-α, had little effect on DV replication and that furthermore in DV-infected cells the normal cellular signaling response to TNF-α was altered.
MATERIALS AND METHODS
Cell lines and culture.
Vero cells and human hepatoma Huh7 and HepG2 cells (American Type Culture Collection) were used for DV infection studies, and L929 mouse fibroblast cells (American Type Culture Collection) were used in the TNF-α bioassay. Vero, HepG2, and Huh7 cells were cultured in Dulbecco modified Eagle medium (DMEM; Gibco), and L929 cells were cultured in RPMI (Gibco) supplemented with 10% fetal bovine serum (FBS) (JRH Biosciences), 1.2 μg/ml penicillin, 1.6 μg/ml gentamicin, 2 mM l-glutamine, and 10 mM HEPES.
Culture and infection of MDM.
Monocytes from healthy donors were isolated by adherence as previously described (50) and cultured for 5 days in complete DMEM containing 10% FBS, 7.5% human serum, 1.2 μg/ml penicillin, 1.6 μg/ml gentamicin, 2 mM l-glutamine, and 10 mM HEPES. On the fifth day after isolation, MDM were detached, seeded in 48-well plates at a concentration of 2 × 105 cells per well, and allowed to readhere. This method of MDM isolation has been previously shown to yield cells that are 85 to 90% CD14 positive (50). The cell population is heterogeneous but shows adherent macrophage-like morphology and has phagocytic activity. Prior to infection, MDM were washed and then infected with MON601 (a laboratory clone of the DV strain New Guinea C [21, 50]) at a multiplicity of infection (MOI) of 5 in a volume of 100 μl for 90 min as previously described (50). Mock-infected control assays were performed by infecting cells, as described above, with equal volumes of heat-inactivated virus (80°C for 20 min). After infection, the cells were washed and cultured in complete DMEM, and one-half the volume of fresh medium was replaced at each sample point.
Quantitation of infectious virus by plaque assay.
Cell culture supernatants from infected cells were assayed for infectious DV by plaque assay. Vero cells were cultured at 3 × 105 cells per well in six-well plates. Cells were washed and infected with 300 μl of serially diluted (10−1 to 10−6) supernatant as described above. Following infection, inoculum was removed and cells were washed and overlaid with 4 ml of a 1:1 mix of 2% Metaphore agarose (BioWhittaker Molecular Applications) and 2× DMEM plus 10% FBS. Agarose was allowed to set, and plates were inverted and incubated at 37°C for 5 days. On day 5, cells were overlaid with 2 ml of 2× DMEM plus 10% FBS containing 0.03% neutral red (MP Biomedicals) and 2% Metaphore agarose and incubated till plaques were visible. Levels of infectious virus were quantitated as PFU per ml. The reproducibility of the plaque assay results was determined by multiple measurement of the same sample (n = 3) in two different experiments, yielding 5 and 10% variation from the mean, respectively, and routinely detected >50 PFU/ml.
Generation of DV capsid construct and in vitro-transcribed RNA.
The DV2 capsid gene was PCR amplified from full-length, infectious clone MON601 with primers CAPECOR1 (5′ CACGAATTC/AGCTCAACGTAGTTCTAACAG 3′) and CAPBAMHI (5′ CGTGGATCC/GATCATGTGTGGTTCTCCGTT 3′) and cloned into pGEM-3Zf(−) (Promega), which contained a T7 forward promoter and an SP6 reverse promoter. Cloning was performed by Robyn Taylor, University of Adelaide. For generation of positive-strand RNA, the pGEM-DV2 capsid was linearized with HindIII and in vitro transcribed with T7 RNA polymerase. For generation of negative-strand RNA, the pGEM-DV2 capsid was linearized with EcoRI and in vitro transcribed with SP6 RNA polymerase. Both in vitro transcription reactions utilized Ambion Maxiscript following the manufacturer's instructions. The in vitro-transcribed RNA was purified using an RNeasy RNA extraction kit (QIAGEN) and quantified by spectrophotometry, and the copy number was calculated.
RNA extraction, tagged reverse transcription (RT), and real-time PCR for viral RNA quantification.
Total RNA was isolated from DV-infected cells using TRIzol (Invitrogen). The RNA was DNase treated by being resuspended in 2 units of RNase-free DNase I (Roche), 10 U RNase inhibitor (Roche), 0.1 M sodium acetate, and 5 mM MgSO4 (pH 5); incubated for 15 min at 37°C; and then phenol-chloroform (BDH) extracted, ethanol precipitated, and resuspended in RNase-free water with 10 U RNase inhibitor. The isolated RNA was reverse transcribed and tagged as follows: RNA was denatured at 65°C for 3 min in the presence of 20 pmol of DV-specific primer attached to a 19-mer sequence, (TAG)-5′ CGGTCATGGTGGCGAATAA 3′, as described in the work of Peyrefitte et al. (49). The primer sequence for the DV positive-strand RNA was TAG-(DV3.2) 5′ ′TAG'TTGTCAGCTGTTGTACAGTCG 3′ and for the DV negative-strand RNA was TAG-(DV5.1) 5′ ′TAG'GCAGATCTCTGATGAATAACCAAC 3′. Ten microliters of denatured RNA (approximately 100 ng) was added to an RT mixture containing 10 U Moloney murine leukemia virus (Biolabs NEB; Genesearch), 10 U RNase inhibitor, 0.5 mM (each) deoxynucleoside triphosphates (Promega) in 1× Moloney murine leukemia virus buffer (Biolabs NEB; Genesearch), and RNase-free water up to 20 μl. Known amounts of negative- and positive-strand in vitro-transcribed DV RNA were reverse transcribed in parallel with the extracted RNA from infected cells to estimate RNA copies in the samples, and extensive controls which included no RT enzyme, no primer, and the wrong primer were used to control for any nonspecific primed cDNA. RT reactions were performed at 37°C for 1 h followed by 95°C denaturation. Two microliters of (1:100) diluted cDNA sample was used in a real-time PCR mixture containing 1× Quantitect SYBR green (QIAGEN) and 0.5 μM of each primer. The DNA primer pair for positive-strand RNA was primers TAG and DV5.1, and that for the negative strand was TAG and DV3.2. Real-time PCRs were performed in a Rotor Gene 3000 real-time thermal cycling system (Corbet Research) for 35 cycles (95°C, 20 s; 58°C, 20 s; 72°C, 20 s). The RT real-time PCR was normalized by determining cyclophilin RNA in total RNA extracted from cells. This involved RT conditions as described above with 0.5 μg oligo(dT)15 (Promega) used as primer for cDNA synthesis. RT real-time PCR was performed using primers CycA(f) (5′ GGCAAATGCTGGACCCAACACAAA 3′) and CycA(r) (5′ CTAGGCATGGGAGGGAACAAGGAA 3′), in the same way as for DV except that 40 cycles were involved (94°C, 20 s; 60°C, 20 s; 72°C, 30 s). Known concentrations of total RNA were used as standards. Cyclophilin mRNA levels did not vary over a 3-day period in DV-infected and uninfected macrophages (data not shown). The sensitivity of DV negative- and positive-strand PCR is 60 to 70 copies/ng cyclophilin mRNA.
Treatment of cells with cytokines.
HepG2 cells were seeded at 2 × 105 cells per well in a 12-well plate, and adherent MDM were seeded at 2 × 105 cells per well in a 48-well plate. To pretreat cells with alpha interferon (IFN-α) or TNF-α, the medium was removed and the required amount of cytokine was added in medium containing 2% FBS and incubated for 4 or 24 h prior to infection. IFN-α (Intron A; Schering-Plough Ltd.) (gift from Karla Helbig, Hepatitis C Lab, University of Adelaide) was used at 100 IU/ml and TNF-α (ProSec-TechnoGene Ltd.) was used at 500 ng/ml. Cells were washed and infected with DV (MON601) at an MOI of 5 as described above. The cells were then washed and resuspended in fresh medium containing 2% FBS (designated as time zero). For posttreatment of cells with IFN-α or TNF-α, the cells were infected as described above and cultured for 24 h, medium was removed, and the required amount of cytokine as described above was added in medium containing 2% FBS and incubated for a further 24 h. At 48 h postinfection (p.i.) supernatant was collected and assayed for infectious virus production (PFU/ml) and cell lysates were analyzed for DV RNA.
Blocking TNF-α using TNF-α antibodies.
Adherent MDM were seeded at 2 × 105 cells per well in 48-well plates and infected with DV at an MOI of 5, as described above. Twenty-four hours later all the supernatant from the wells was replaced with medium containing either 2.5-μg/ml purified mouse anti-human TNF-α antibody (catalog no. 18630D; Pharmingen) or the recommended isotype-matched control (catalog no. 20800D; Pharmingen) or medium without any additive. At 48 h p.i. all the supernatant was removed from the wells and replaced with fresh medium, and at 72 h p.i. supernatant was collected and assayed for virus release (PFU/ml) and total RNA was extracted to analyze viral RNA production in cells.
Transfection of siRNAs into MDM.
Predesigned Stealth duplex oligonucleotide short interfering RNAs (siRNAs) were obtained from Invitrogen. The sequences of the sense and antisense strands were as follows: TNF-HSS110854 (siRNA 4), 5′ UCGAGAAGAUGAUCUGACUGCCUGG 3′ and 5′ CCAGGCAGUCAGAUCAUCUUCUCGA 3′; HSS110855 (siRNA 5), 5′ UUAUCUCUCAGCUCCACGCCAUUGG 3′ and 5′ CCAAUGGCGUGGAGCUGAGAGAUAA 3′; HSS110856 (siRNA 6), 5′ AGACUCGGCAAAGUCGAGAUAGUCG 3′ and 5′ CGACUAUCUCGACUUUGCCGAGUCU 3′. The negative-control siRNA used was Stealth RNA interference negative-control duplex “medium GC content” (Invitrogen). siRNA transfection efficiency was determined by transfecting Block-iT fluorescent oligonucleotide (Invitrogen) using Lipofectamine (Invitrogen).
The day before transfection, adherent MDM were seeded at concentrations of 2 × 105 cells per well in a 48-well plate in MDM growth medium without antibiotics. Cationic lipid complexes were prepared by incubating 1.6 μM siRNA duplexes with 0.6 μl Lipofectamine RNAiMAX (Invitrogen) in 40 μl of DMEM without serum and then added to the cells. After 4 h of incubation, the cells were washed and replaced with fresh 200 μl of MDM growth medium without antibiotics for at least 18 h to allow recovery, followed by a second round of siRNA transfection as described above. After 4 h of incubation the cells were washed and resuspended in MDM growth medium.
DV infection and LPS stimulation of siRNA-transfected cells.
MDM were transfected with TNF-α siRNA as described above, and 18 h after transfection the cells were infected with DV at an MOI of 5. At 48 h p.i. supernatants were collected, to measure infectious virus release by plaque assay and TNF-α production. Intracellular RNA was also extracted to monitor DV RNA synthesis. To validate the efficacy of siRNA knockdown of TNF-α protein, TNF-α production was stimulated by addition of 10 ng/ml lipopolysaccharide (LPS; Sigma) for 4 h to siRNA-transfected cells. Supernatant was collected, and cells were thoroughly washed using Hanks balanced salt solution (Gibco) and refreshed with MDM growth medium. LPS exposure (as described above) was repeated daily up to day 4 posttransfection. The supernatants were assayed for TNF-α bioactivity using an L929 cytotoxic bioassay (see below). Additionally, TNF-α protein release from DV-infected cells, with or without siRNA transfection, was quantitated by an enzyme-linked immunosorbent assay (ELISA; Pharmingen; OptEIA human TNF-α set) in accordance with the manufacturer's recommendation.
L929 cytotoxic bioassay.
L929 cytotoxicity was analyzed by modifying the method from reference 14. L929 cells were plated in 96-well plates at a density of 104 cells per well and incubated overnight. Known concentrations of TNF-α (1,000 pg/ml) (used as standards) and cell supernatants (see above) were serially diluted 10-fold in RPMI medium containing 2% FBS and actinomycin D (Sigma) at 2 μg/ml, and 100 μl of TNF-α standards or diluted samples was added to the cells. The cells were reincubated for 18 h and fixed with 100% methanol, and viable cells were stained using 0.5% crystal violet (BDH) diluted in 25% methanol. After 5 min of staining the monolayers were washed extensively with water, and dye adherent to plates was solubilized using 1% sodium dodecyl sulfate (BDH). Absorbance was read at 540 nm, standards were graphed, and concentrations of TNF-α in samples were determined. The specificity of this assay for TNF-α was shown by blocking bioactivity of TNF-α by coincubation of LPS-stimulated supernatants from TNF-α siRNA-transfected macrophages and 50,000 pg/ml of TNF-α with 2.5 μg/ml TNF-α monoclonal antibodies for 2 h prior to the L929 cytotoxicity assay.
Quantitation of TNF-α-stimulated NF-κB LUC reporter gene transcription.
Huh7 cells were seeded at a concentration of 1.4 × 105 cells per well in a six-well plate and then cotransfected with two different reporter plasmids (2 μg each) and 10 μl Superfect (QIAGEN) per the manufacturer's instructions. The first plasmid, pT81-NFκB-LUC (a gift from Andrew Bert, Human Immunology, Institute of Medical and Veterinary Science, Adelaide, Australia), was a modified version of pT81-LUC (45) and contained five tandem repeats of a 27-bp NF-κB-responsive element derived from the immunoglobulin κ gene enhancer (aacagagGGGACTTTCCgaggccatct, with the NF-κB binding site in capital letters) upstream of the thymidine kinase (TK) promoter. The second plasmid, pRL-TK (Promega), contained a Renilla luciferase (LUC) gene under the control of a constitutive TK promoter. After 18 h, the transfected cells were trypsinized, distributed equally into four wells of a 24-well plate, and allowed to adhere for 18 h. Duplicate wells were infected with DV at an MOI of 5 or mock infected as described above. Twenty-four hours p.i., 10 ng/ml of TNF-α or fresh medium alone was added and incubated for 6 h before the cells were harvested and lysed. LUC activity was quantitated using Promega's Dual Luciferase Reporter Assay System Kit. The activity of firefly LUC was normalized to the activity of Renilla LUC or to the total protein concentration measured in a Bio-Rad Dc protein assay system. The two methods of normalization yielded the same result. As a control the IFN-α-responsive plasmid pISRE-LUC (Clontech Labs) was also transfected into cells, the transfected cells were stimulated with 100 IU IFN-α, and the LUC activity was measured and normalized as described above.
Analysis of TNF-α-stimulated NF-κB movement using confocal microscopy.
MDM and Huh7 cells were seeded at 1 × 105 and 1 × 104 cells per well, respectively, in a 16-well Lab-Tek chamber glass slide (Nunc) and infected at an MOI of 5. MDM at 2 days p.i. or Huh7 cells at 1 day p.i. were treated with TNF-α (10 ng/ml) for 30 min or not treated. Cells were fixed in 1% formalin and permeabilized with 0.05% IGEPAL CA-630 (Sigma). MDM were then blocked with 4% human serum, 4% goat serum, and 0.4% bovine serum albumin. Huh7 cells were blocked with 4% goat serum and 0.4% bovine serum albumin diluted in Hanks balanced salt solution containing 5% fetal calf serum and 0.02% sodium azide (Ajax Chemicals). After 30 min of blocking, the MDM were incubated with mouse monoclonal anti-DV E antibodies (a gift from Peter Wright, Department of Microbiology, Monash University, Clayton, Victoria, Australia) (1/50) and rabbit anti-NF-κB p-65 (a gift from Michael Beard, Hepatitis C Lab, University of Adelaide, Australia) (1/50). Huh7 cells were incubated with DV-positive patient sera (1/5,000) and rabbit NF-κB anti-p-65 (1/50). Following 45 min of incubation the MDM were incubated with a 1/200 dilution of goat anti-mouse Alexa Fluor 546 (Molecular Probes) and goat anti-rabbit Alexa Fluor 488, while the Huh7 cells were incubated with a 1/200 dilution of goat anti-human Alexa Fluor 546 and goat anti-rabbit Alexa Fluor 488. Cells were mounted onto glass slides with ProLong antifade mounting medium (Molecular Probes) and examined by confocal microscopy. Confocal images were captured at ×400 magnification using a Bio-Rad Radiance 2100 confocal microscope, and fluorescent intensities were adjusted using Confocal Assistant (Bio-Rad). The specificity of staining was shown using uninfected cells stained with DV antibodies and DV-positive cells stained with a species-matched irrelevant antiserum.
RESULTS
Characteristics of DV replication in MDM.
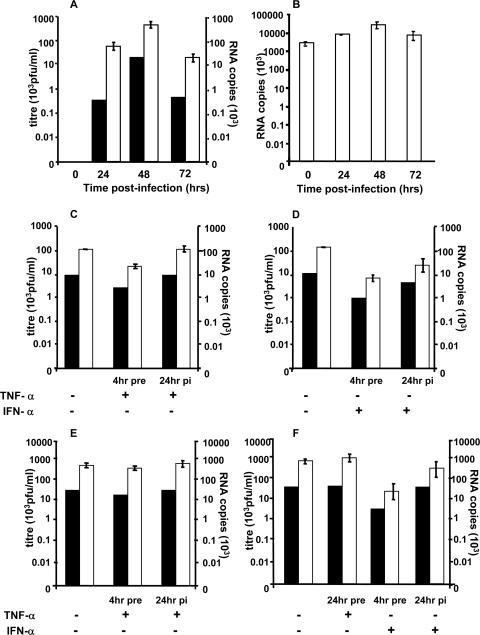
The time course of DV replication over 72 h was determined. Adherent MDM, 6 days after isolation, were infected, and at different time points after infection, supernatant was collected to quantitate the release of infectious virus (PFU/ml) and intracellular RNA was extracted from cell lysates to measure levels of positive- and negative-strand DV RNA. A tagged RT real-time PCR was used to minimize self-priming of DV RNA that has been observed to occur in our laboratory and reported by others during the RT step (49). Active replication of DV in MDM was shown by production of infectious virus (PFU/ml) and increasing levels of positive- and negative-strand RNA, with maximum virus and RNA levels at 48 h p.i. and levels of virus and RNA production which continued at a lower level on day 3 (Fig. 1A and B). The de novo production of negative-strand RNA coincided with infectious virus production, whereas positive-strand RNA was present at high levels at all time points with much smaller relative changes associated with infectious virus production. Confocal microscopy of DV-infected cells stained for DV antigens showed that 15 to 30% of MDM were infected at 48 h p.i., the time of peak virus production (data not shown). DV-positive MDM, visualized by immunofluorescence (see Fig. 6), were adherent and had heterogeneous but macrophage-like morphology and the ability to phagocytose sheep red blood cells (data not shown). Infection of MDM by DV showed no visual cytopathic effects. Subsequent studies utilized this infection model and monitored DV replication by release of virus (plaque assay) and accumulation of negative-strand RNA (RT-PCR).
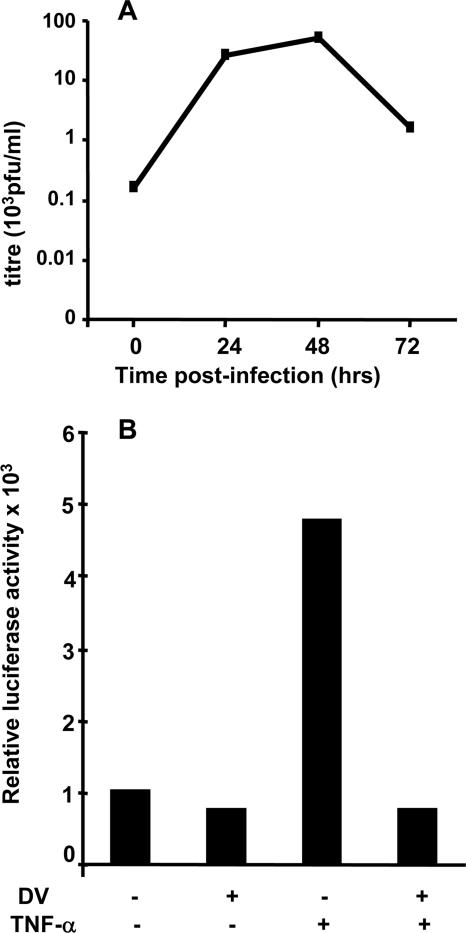
FIG. 1.
Exogenously added TNF-α has no major effect on established DV replication in MDM but exerts a minor effect when added 4 h prior to DV infection. (A and B) DV replication in MDM. MDM were infected with DV at an MOI of 5, supernatant was collected at various time points p.i., and RNA was extracted from cell lysates. Intracellular DV RNA was quantitated using a tagged RT real-time PCR. In vitro-transcribed copy number standards from DV2 capsid protein were used to quantitate DV RNA. Tagged RT real-time PCR results represent averages ± standard errors of the means (n = 2) in the PCR assay and were normalized per ng cyclophilin RNA. Black bars, viral titer; open bars, DV RNA. (A) Infectious DV release (plaque assay, PFU/ml) and negative-strand DV RNA. (B) Positive-strand DV RNA. (C to F) Effect of TNF-α. MDM and HepG2 cells were exposed to TNF-α or IFN-α for 4 or 24 h and then infected with DV at an MOI of 5 or infected and then cytokine treated 24 h p.i. At 48 h p.i. supernatants were collected for viral plaque assays, and RNA was extracted for analysis of DV RNA, as in panels A and B. Values represent means ± standard errors of the means from duplicate samples in RT real-time PCR of a representative experiment. Experiments were replicated (n = 2, RNA; n = 4, titer). (C) TNF-α in MDM; (D) IFN-α controls in MDM; (E) TNF-α in HepG2 cells; (F) TNF-α and IFN-α controls in HepG2 cells.
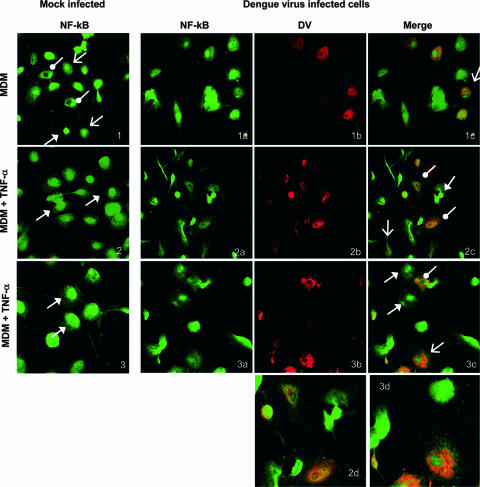
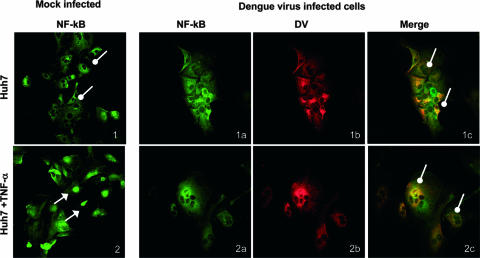
FIG. 6.
DV inhibits TNF-α-stimulated translocation of NF-κB into the nucleus in 50% of DV-positive MDM. MDM were mock infected with heat-inactivated virus or infected with DV and treated with TNF-α or not treated. After 30 min cells were fixed and stained with antibodies to DV E antigen (red Alexa Fluor 546) and p65 NF-κB (green Alexa Fluor 488) and visualized by confocal microscopy. White arrows with solid heads, nuclear NF-κB; white lollipops, cytoplasmic NF-κB proteins; white arrows with open heads, NF-κB proteins in both nucleus and cytoplasm. Panel 1, mock-infected MDM not treated with TNF-α; panels 2 and 3, mock-infected MDM treated with TNF-α; panels 1a to c, DV-infected MDM not treated with TNF-α; panels 2a to c and 3a to c, DV-infected MDM treated with TNF-α; panels 2d and 3d, magnified images from panels 2c and 3c, respectively (DV-infected MDM treated with TNF-α). Of MDM, 24 to 30% were DV positive by comparison with uninfected controls stained for DV antigens or DV-infected MDM stained with a matched control, normal mouse serum (data not shown). Results are representative of five fields (average, 10 cells per field) viewed from each of three independent experiments.
Addition of exogenous TNF-α prior to or after established infection does not dramatically affect DV replication in MDM.
TNF-α is present at high levels in the circulation of DV-infected patients and is released by DV-infected macrophages and other cells normally present in the environment of macrophages. Pretreatment of HepG2 cells with IFN-α 4 h before infection inhibits subsequent DV replication while pretreatment with TNF-α for 24 h before infection has no effect on DV replication in HepG2 cells (15) and MDM (10). Prior studies in our laboratory have shown that TNF-α is released by DV-infected MDM, coinciding with the peak of DV production at 36 h after infection (7). We therefore looked at the effect on DV replication of adding TNF-α to MDM, both at 4 h prior to infection and at 24 h p.i., a time point that represents established infection but is prior to the peak of endogenous TNF-α release and progeny virus production. HepG2 cells pretreated and posttreated before infection with TNF-α and IFN-α were used as comparative controls (15). The level of TNF-α used in this part of our study was higher than the pg/ml levels released from DV-infected MDM (7), consistent with levels known to induce TNF-α responses in the literature (32, 41), and higher than the levels used later in our study to induce TNF-α signaling in uninfected cells and thus should be in the required range for TNF-α activity.
Analysis of levels of DV RNA and infectious virus at 48 h p.i. showed no effect of addition of TNF-α at 24 h p.i. on subsequent DV replication in MDM (Fig. 1C) or HepG2 cells (Fig. 1E). In contrast, treatment with IFN-α at 24 h p.i. showed a moderate inhibitory effect on DV negative-strand RNA accumulation and virus release from MDM (Fig. 1D) and, as previously reported, also in HepG2 cells (Fig. 1F) (15).
Pretreatment of MDM with TNF-α 4 h before DV infection had a small reproducible inhibitory effect (two- to fourfold) on DV replication (Fig. 1C). The paired Student t test from the results of four independent experiments measuring infectious virus release (PFU/ml) and two independent experiments that quantitated RNA levels showed that, although alteration in the release of infectious virus was not significant (P = 0.28), there was a significant reduction in RNA levels (P = 0.03), suggesting this to be a real and reproducible inhibition of DV replication. However, pretreatment of HepG2 cells for 4 h with TNF-α had no effect (<2-fold) on DV replication (Fig. 1E). These moderate effects of pretreatment with TNF-α contrast with the more significant effects seen with IFN-α, where 4 h of pretreatment of MDM and HepG2 cells with IFN-α showed a >10-fold inhibition of negative-strand RNA accumulation and virus release (Fig. 1D and F), consistent with the literature (15). Finally, we saw no effect of 24-h TNF-α pretreatment on DV replication in HepG2 cells (Fig. 1E) and MDM (results not shown), again consistent with previous reports (10, 15). Thus, addition of high levels of exogenous TNF-α, which is known to be stable in culture conditions (1), had no major effect on established DV replication in MDM and had a minor inhibition of DV replication in MDM only if added 4 h prior to infection. The small inhibition of DV replication in MDM following a short (4-h) but not longer (24-h) pretreatment with TNF-α may imply that TNF-α stimulates short-lived antiviral responses, and further investigation may be of interest in understanding the mechanisms by which TNF-α can induce an antiviral state in uninfected cells and the timing of such effects.
Inhibition of endogenously produced TNF-α has no effect on DV replication in MDM.
Since DV-infected macrophages can produce their own TNF-α, we investigated if using siRNA to block endogenous TNF-α produced by infected cells or using TNF-α antibodies to block actions of TNF-α had any effect on DV replication in these cells.
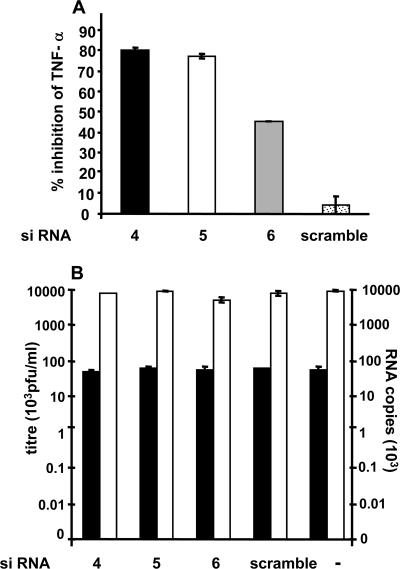
To achieve knockdown of production of endogenous TNF-α, three predesigned stealth TNF-α siRNAs (Invitrogen) were used to transfect MDM. Stealth siRNAs were used to prevent stimulation of the double-stranded RNA response that could complicate the interpretation of results. Establishment of siRNA transfection conditions using a fluorescent siRNA (Invitrogen) showed >90% uptake of the labeled oligonucleotides into MDM (data not shown). We next validated TNF-α protein knockdown. One day after siRNA transfection, MDM were stimulated with 10 ng/ml of LPS and TNF-α release was measured by bioassay. Results showed 50 to 80% inhibition of bioactive TNF-α protein release mediated by the three siRNAs over a 4-day period (data not shown). This indicates rapid and persistent TNF-α knockdown in MDM from 1 to 4 days posttransfection using these siRNA transfection conditions.
To test if siRNA reduction of TNF-α in MDM had any effect on DV replication, we transfected MDM with the same three stealth TNF-α siRNAs and a negative control (scramble siRNA) and then infected them with DV (MOI of 5). Experiments were sampled at 32 h p.i., since at the usual sampling point at 48 h, TNF-α mRNA could not be detected to confirm siRNA mRNA knockdown. We know, however, that DV RNA and infectious virus levels are similar at these two time points. At 32 h p.i. supernatant was harvested and analyzed for viral titer, DV RNA, and TNF-α mRNA and protein production. Two of the three stealth siRNAs showed >75% inhibition of TNF-α mRNA (data not shown) and protein release from DV-infected MDM (Fig. 2A), showing efficient knockdown of endogenous TNF-α. However, even with this high level of TNF-α protein knockdown we still observed no difference in virus replication (Fig. 2B). The lack of effect of siRNA knockdown of TNF-α on DV replication was also observed at 48 h p.i., although concomitant siRNA knockdown of TNF-α could not be confirmed in these latter experiments.
FIG. 2.
siRNA knockdown of TNF-α in MDM has no effect on DV replication. MDM were transfected with individual TNF-α siRNAs by two rounds of 4-h transfection with 1.6 μM TNF-α siRNA 4, 5, or 6 infected with DV at an MOI of 5, and at 32 h p.i. cell culture supernatants were analyzed. (A) TNF-α production (ELISA). Results were expressed as a percentage of TNF-α released from DV-infected mock-transfected MDM (2,027 pg/ml/24 h). Values are means ± standard errors of the means of two wells. (B) DV replication determined by infectious virus release (PFU/ml, black bars) and negative-strand RNA levels (white bars) in infected cells. In vitro-transcribed copy number standards from DV2 capsid protein were used to quantitate negative viral RNA using a tagged RT real-time PCR. Values represent means ± standard errors of the means from duplicate samples in RT real-time PCR of a representative experiment per ng cyclophilin RNA. Experiments were replicated (n = 2).
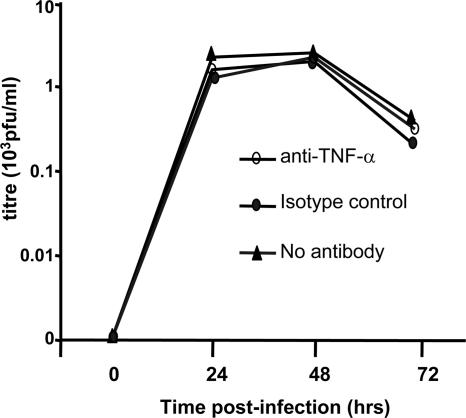
Although we achieved high-level TNF-α mRNA and protein knockdown, TNF-α was not completely abolished. We thus next aimed to extend the siRNA knockdown studies by antibody blocking of endogenous TNF-α activity. MDM were isolated and infected with DV, and at 24 h p.i., prior to the peak of TNF-α release (7), neutralizing TNF-α antibody at 2.5 μg/ml or isotype-matched control antibodies were added. This amount of TNF-α antibody is capable in our laboratory of neutralizing >50,000 pg/ml of TNF-α activity by bioassay (data not shown). In comparison, the cumulative level of TNF-α released from DV-infected MDM in this study at 32 to 48 h p.i. was 1,980 to 2,185 pg/ml, as determined by TNF-α ELISA (data not shown). Cultures were supplemented with fresh medium and antibodies throughout the experiment. DV replication profiles in the presence and those in the absence of TNF-α antibodies showed little difference, suggesting that endogenously released TNF-α has no effect on DV replication in MDM (Fig. 3). Further, cellular RNA extracted at 72 h showed no difference in levels of DV RNA (data not shown).
FIG. 3.
Antibody blocking of endogenous TNF-α had no effect on DV replication. MDM were infected with DV at an MOI of 5 and 24 h after infection cells were treated with either anti-TNF-α antibodies (2.5 μg/ml), isotype-matched control (2.5 μg/ml), or fresh medium. Supernatant was collected every 24 h and replaced with fresh antibody or medium. Infectious virus release was detected by plaque assay (PFU/ml). Results are from a representative experiment and were replicated (n = 3). No significant differences were observed in any experiment.
DV-infected cells do not respond normally to exogenously added TNF-α.
Infection with hepatitis C virus, which like DV belongs to the Flaviviridae family, has been shown to confer resistance to TNF-α and alter TNF-α signaling in infected cells (13, 17, 40, 47, 54). Since exogenous and endogenously added TNF-α had no effect on DV replication, we aimed to investigate if DV-infected cells could still respond to TNF-α by analysis of TNF-α-induced, NF-κB promoter-directed transcription, and NF-κB nuclear translocation.
In contrast to the efficient uptake of siRNA observed in MDM, large DNA constructs transfect these cells poorly. Thus, we used Huh7 hepatoma cells for these studies. Huh7 cells support active DV replication as shown by production of infectious virus with time (Fig. 4A). DV infection was cytolytic, resulting in cell death by 48 to 72 h p.i. (data not shown), and thus, all analysis was performed at 24 h p.i. when there is active replication and approximately 100% of cells were infected as shown by immunostaining without a high degree of cell death (results not shown). Huh7 cells were cotransfected with two plasmids: (i) pT81-NF-κB-LUC, which regulates the transcription of firefly LUC under an NF-κB-responsive promoter, and (ii) pRL-TK, which contains the Renilla LUC gene under the control of a constitutive TK promoter. Transfected cells were infected with DV or mock infected with heat-inactivated virus, and 24 h p.i., cells were either left untreated or treated with TNF-α and LUC activity was measured (Fig. 4B). Mock-infected, untreated cells showed basal levels of NF-κB-mediated reporter gene transcription, and DV-infected cells without TNF-α treatment had similar levels of LUC activity (Fig. 4B). Addition of TNF-α to mock-infected cells saw a fivefold increase in LUC activity, consistent with TNF-α stimulation of NF-κB-mediated reporter gene transcription (Fig. 4B). In contrast, TNF-α stimulation of DV-infected cells failed to show any increase in LUC activity. This lack of response to TNF-α was similar in magnitude to DV inhibition of IFN-α signaling (15; data not shown). Similar results were obtained from experiments performed in 293-T cells, although in this cell type a more modest 25 to 30% reduction in TNF-α-stimulated LUC reporter gene activity in DV-infected cells was observed (data not shown).
FIG. 4.
DV-infected Huh7 cells do not respond to TNF-α stimulation. (A) Replication profile of DV in Huh7 cells. Huh7 cells were infected with DV at an MOI of 5, and supernatant was collected and assayed for infectious virus release (PFU/ml). The peak of virus production was seen at 48 h p.i. (B) DV inhibits TNF-α-stimulated NF-κB-responsive transcription. Huh7 cells were cotransfected with plasmids, containing the firefly LUC gene downstream of an NF-κB-responsive promoter. At 24 h p.i. cells were treated with TNF-α (10 ng/ml) for 6 h or not treated. Cells were lysed and normalized against protein concentration, and reporter activity was measured. Results shown are representative of three independent experiments.
Since the reporter assays shown in Fig. 4B cannot be performed in primary MDM, we investigated the effects of TNF-α stimulation of DV-infected MDM by analyzing TNF-α-induced NF-κB nuclear translocation by immunostaining for p65 NF-κB proteins and confocal microscopy and used Huh7 cells for comparison. DV-infected and mock-infected Huh7 cells that were not treated with TNF-α showed cytoplasmic staining of NF-κB p65 proteins as expected (Fig. 5, panel 1). TNF-α stimulation of mock-infected Huh7 cells led to clear translocation of p65 NF-κB proteins to the nucleus in >95% of cells (Fig. 5, panel 2; Table 1). However, TNF-α stimulation of DV-infected Huh7 cells failed to induce nuclear localization of NF-κB (Fig. 5, panel 2a to c) in cells that were confirmed to be DV infected (Fig. 5, panel 2b). These data support the lack of TNF-α signaling in DV-infected cells seen in reporter assays shown in Fig. 4B. Similar results were observed in DV-infected HepG2 cells but not in the lung fibroblast cell line A549, which, unlike DV target cells such as MDM, Huh7, and HepG2 liver cells, undergoes rapid and complete DV-induced cell death within 48 h (data not shown). Cell-type-specific blocking of TNF-α signaling by DV and its relationship to DV replication or induction of cell death need to be investigated further.
FIG. 5.
DV inhibits TNF-α-stimulated translocation of NF-κB proteins into the nucleus in Huh7 cells. Cells were mock infected with heat-inactivated virus or infected with DV and treated with TNF-α for 30 min or not treated. Cells were fixed and stained with DV-positive patient serum (red Alexa Fluor 546) and p65 NF-κB (green Alexa Fluor 488) and visualized by confocal microscopy. White arrows, nuclear NF-κB; white lollipops, cytoplasmic NF-κB. Panels 1 and 2, mock-infected Huh7 cells not treated or treated with TNF-α, respectively; panels a to c, DV-infected Huh7 cells. More than 95% of Huh7 cells were DV positive by comparison with uninfected controls stained for DV antigens or DV-infected Huh7 cells stained with a control DV-negative human serum (data not shown).
TABLE 1.
Quantitation of NF-κB cellular localization after TNF-α treatment or no TNF-α treatment
| Treatment and cell type | % of cells localized to:
|
|||||
|---|---|---|---|---|---|---|
| Nucleus
|
Cytoplasm
|
Nucleus and cytoplasm
|
||||
| + TNF-α | − TNF-α | + TNF-α | − TNF-α | + TNF-α | − TNF-α | |
| Huh7 | ||||||
| Mock | 100 | 0 | 0 | 100 | ||
| DV | 0 | 0 | 100 | 100 | ||
| MDMa | ||||||
| Mock | 100 | 48 | 0 | 2 | 0 | 50 |
| DV positiveb | 4 | 10 | 48 | 8 | 48 | 82 |
| DV negativeb | 100 | 48 | 0 | 2 | 0 | 50 |
Results for MDM are representative of five fields (average, 10 cells per field) viewed from each of three independent experiments.
As assessed by DV antigen staining in the DV-challenged MDM population.
In contrast to the clear cytoplasmic staining for NF-κB in untreated Huh7 cells, mock-infected MDM that were not treated with TNF-α showed a variable distribution of intracellular NF-κB p65 proteins with most MDM showing NF-κB proteins in both cytoplasm and nucleus or the nucleus exclusively (Fig. 6, panel 1; Table 1). Similarly, DV-infected cells without TNF-α treatment showed NF-κB proteins in both cytoplasm and nucleus or in the nucleus alone in most cells (Fig. 6, panels 1a to 1c). However, after stimulation with TNF-α, mock-infected MDM clearly showed nuclear translocation of NF-κB p65 protein in 100% of the cells (Fig. 6, panels 2 and 3). In contrast dual analysis for NF-κB and DV antigens in DV-infected MDM showed TNF-α stimulation of NF-κB nuclear translocation in most DV-negative but only some DV-positive cells within the MDM population (Table 1). Conversely, the remaining 50% of TNF-α-stimulated, DV-positive staining MDM showed no nuclear NF-κB (Fig. 6, panels 2 and 3a to 3c). The localization of NF-κB to the cytoplasm alone after TNF-α stimulation was observed only in DV-infected cells. Magnified images are shown for clarity (Fig. 6, panels 2 and 3d). These results suggest that the lack of nuclear movement of NF-κB in response to TNF-α stimulation occurs only in DV-infected cells but that effective TNF-α induction of NF-κB nuclear translocation can still occur in uninfected bystander cells and some DV-infected MDM.
Together, the observed lack of nuclear translocation of NF-κB in response to TNF-α in DV-infected Huh7 and HepG2 cells and reduced nuclear movement of NF-κB in DV-infected MDM confirm the reporter assays shown in Huh7 cells and suggest that during established DV infection of relevant target cell types such as macrophages and liver cells, DV-infected cells are unable to respond normally to TNF-α stimulation.
DISCUSSION
Our results show that DV replication in a relevant primary cell system, MDM, which represent a key target of DV infection in vivo (24, 53), is largely unaffected by TNF-α and further show an altered ability of DV-infected MDM and hepatoma cells to effectively respond to TNF-α stimulation.
Addition of high concentrations of TNF-α had little effect on DV replication with only a very small inhibition of DV replication in macrophages following 4-h pretreatment with TNF-α. Similarly, blocking the activity of endogenously produced TNF-α by siRNA knockdown or incubation of DV-infected MDM with TNF-α antibodies also had no effect on DV replication. These results strongly suggest that DV replication is not dramatically affected by either the addition of exogenous or the blocking of endogenous TNF-α in the natural target cell, the macrophage. With WNV, another flavivirus, addition of TNF-α 1 h p.i. significantly reduces the number of infected cells from TNF-α-deficient mice but has no effect in cells from wild-type animals (11). These results of WNV infection in wild-type cells show some similarities to our results for DV replication with a small effect of pretreatment but lack of effect of exogenously added TNF-α on DV replication when added at later stages of infection. However, the lack of an antiviral action of TNF-α contrasts sharply with the significant antiviral actions of TNF-α seen in other viral systems such as CMV (48) and herpes simplex virus type 1 (50).
Sera from patients with dengue hemorrhagic fever/dengue shock syndrome contain greater levels of several proinflammatory cytokines including TNF-α than do sera from individuals with uncomplicated dengue fever (19, 20, 27, 30, 51), endothelial cells can be activated by culture fluids from peripheral blood monocytes infected with DV which can be abolished by treatment of monocyte culture fluids with TNF-α antibody (2, 6), and anti-TNF-α treatment of mice infected with DV decreases fatality (3, 56), suggesting that TNF-α is at least in part responsible for DV pathogenesis. Anti-TNF-α antibody treatment, however, does not affect DV viremia in vivo (3), consistent with our findings that blocking TNF-α does not affect DV replication in cells in vitro. Thus, measures to block TNF-α actions could improve clinical management of dengue complications, and our studies show that such approaches would not compromise any ongoing antiviral role of TNF-α against DV replication.
There are reports in the literature of hepatitis C virus blockade of TNF-α signaling (13, 40, 47, 54) and DV blockade of IFN signaling (43, 44). Since we found no effect of TNF-α on DV replication, we further investigated the ability of DV-infected cells to respond to TNF-α. TNF-α treatment of uninfected Huh7 cells stimulated NF-κB nuclear translocation and reporter gene transcription, but TNF-α treatment of DV-infected Huh7 cells did not. In MDM, addition of TNF-α to mock-treated cells resulted in nuclear movement of NF-κB in all cells. In stark contrast, addition of TNF-α to DV-infected MDM resulted in movement of NF-κB in DV-negative staining cells but a lack of NF-κB nuclear translocation in approximately 50% of DV-positive-staining cells within the DV-challenged MDM population. Together these results clearly suggests DV inhibition of TNF-α-stimulated NF-κB activation in Huh7 cells and a significant proportion of DV-infected MDM. The observation that only 50% of DV-infected MDM have altered TNF-α responses could be attributed to a number of factors including differences in the stage of macrophage maturation or DV replication, as is seen for NF-κB translocation only during the late stages of infection with CMV (29).
In contrast to our observed DV blockade of TNF-α-stimulated NF-κB nuclear translocation, other studies have shown DV activation of NF-κB (4, 8, 38), including a study showing DV-induced nuclear movement of NF-κB in 60% of DV-infected A549 cells (8). In these studies, including studies with HepG2 hepatoma cells, similar to those used herein, the infection models are immortalized cell lines and investigations were performed at a time at which these cells undergo DV-induced apoptosis, a cell death pathway well documented to involve NF-κB activation. We have performed some limited DV infection studies in A549 cells and observed DV-induced nuclear movement of NF-κB in a much smaller percentage of cells (10%) compared with the results of the work of Chang et al. (8), but within 48 h these DV-infected cells are all killed (data not shown). In our extensive studies reported herein, using in vitro cell models representing DV infection of macrophages and liver cells, two important primary cell targets for DV in vivo, we did not observe DV induction of nuclear NF-κB movement but instead observed DV blockade of TNF-α-stimulated NF-κB activation, at a time of established DV infection but with very little (3 to 10%) apoptosis (16, 42). Thus, while high-level DV infection of cell lines may activate NF-κB and induce apoptosis, during active DV replication in primary MDM or hepatoma cell lines DV does not activate NF-κB and in fact has the capacity to prevent its activation by TNF-α.
This is the first study to show that some DV-infected MDM do not respond normally to TNF-α stimulation. A similar observation has been made for DV-infected dendritic cells (46). TNF-α plays a role in differentiating dendritic cells (by increasing expression of costimulatory and activation molecules), and recent experiments have shown that DV-infected dendritic cells do not up regulate costimulatory molecules CD80 and CD86 in response to TNF-α (46). Thus, DV blocks responses to TNF-α in infected dendritic cells as well as in macrophages (as demonstrated in the current study), which via disruption of the NF-κB signaling pathway may alter the stimulation of many genes normally involved in immune responses. This may complement the previously described DV-induced inhibition of IFN-α signaling and yield multiple mechanisms for subversion of the host immune response. Alternatively, our observed blockade of TNF-α-stimulated NF-κB activation in DV-infected cells may be an indirect consequence of viral mechanisms that prevent NF-κB-mediated apoptosis or of dominant utilization of essential components of the TNF-α signaling pathway by DV that consequently results in the inability of these factors to mediate TNF-α signaling. Further work is needed to determine the mechanism of DV inhibition of responsiveness to TNF-α in order to understand its biological relevance. Inhibition of TNF-α activation of NF-κB in hepatitis C virus infection can be mediated by three hepatitis C virus proteins (core, NS5A, and NS5B) (13, 40, 47, 54), while alteration to expression of IFN-stimulated genes by DV NS4B, NS4A, and NS2A and blockade of STAT-1 and ISRE promoter activation by NS4B (43, 44) suggest a role for these DV proteins in interfering with IFN signaling. Similarly, our current studies aim to determine which DV proteins can mediate inhibition of TNF-α responsiveness in DV-infected cells and whether this is also accompanied by biological changes such as cell susceptibility to apoptosis.
In conclusion, we have demonstrated that DV replication in MDM is largely unaffected by addition of high levels of TNF-α or by blocking production of endogenous TNF-α released by these cells. We also have shown that DV-infected cells do not respond normally to TNF-α stimulation by analyzing transcription from an NF-κB-responsive promoter and viewing NF-κB cellular location. These findings suggest that excessive levels of TNF-α seen in DV-infected patients do not reduce viral replication in DV-infected macrophages and that active DV replication occurs in a cellular state of nonresponsiveness to TNF-α which may reflect a viral mechanism for subverting host antiviral responses or utilizing host cell functions.
Acknowledgments
We thank Karla Helbig and Michael Beard for cells, reagents, and plasmids used in this study; the Australian Red Cross blood bank for buffy coats used to isolate monocytes; Robyn Taylor for plasmids containing DV capsid protein and DV primer sequences; Peter Brautigan for cyclophilin primer sequences; Ghafar Sarvestani for assistance in confocal microscopy; Peta Grant for assistance in the preparation of confocal images for publication; and members of the HIV research lab, Institute of Medical and Veterinary Science.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Aderka, D., H. Engelmann, Y. Maor, C. Brakebusch, and D. Wallach. 1992. Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J. Exp. Med. 175:323-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R., S. Wang, C. Osiowy, and A. C. Issekutz. 1997. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J. Virol. 71:4226-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atrasheuskaya, A., P. Petzelbauer, T. M. Fredeking, and G. Ignatyev. 2003. Anti-TNF antibody treatment reduces mortality in experimental dengue virus infection. FEMS Immunol. Med. Microbiol. 35:33-42. [DOI] [PubMed] [Google Scholar]
- 4.Avirutnan, P., P. Malasit, B. Seliger, S. Bhakdi, and M. Husmann. 1998. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J. Immunol. 161:6338-6346. [PubMed] [Google Scholar]
- 5.Bonner, S. M., and M. A. O'Sullivan. 1998. Endothelial cell monolayers as a model system to investigate dengue shock syndrome. J. Virol. Methods 71:159-167. [DOI] [PubMed] [Google Scholar]
- 6.Cardier, J. E., E. Marino, E. Romano, P. Taylor, F. Liprandi, N. Bosch, and A. L. Rothman. 2005. Proinflammatory factors present in sera from patients with acute dengue infection induce activation and apoptosis of human microvascular endothelial cells: possible role of TNF-alpha in endothelial cell damage in dengue. Cytokine 30:359-365. [DOI] [PubMed] [Google Scholar]
- 7.Carr, J. M., H. Hocking, K. Bunting, P. J. Wright, A. Davidson, J. Gamble, C. J. Burrell, and P. Li. 2003. Supernatants from dengue virus type-2 infected macrophages induce permeability changes in endothelial cell monolayers. J. Med. Virol. 69:521-528. [DOI] [PubMed] [Google Scholar]
- 8.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-kappaB activation. Microbes Infect. 8:157-171. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y. C., and S. Y. Wang. 2002. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J. Virol. 76:9877-9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y. C., S. Y. Wang, and C. C. King. 1999. Bacterial lipopolysaccharide inhibits dengue virus infection of primary human monocytes/macrophages by blockade of virus entry via a CD14-dependent mechanism. J. Virol. 73:2650-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, Y., N. J. King, and A. M. Kesson. 2004. The role of tumor necrosis factor in modulating responses of murine embryo fibroblasts by flavivirus, West Nile. Virology 329:361-370. [DOI] [PubMed] [Google Scholar]
- 12.Chisari, F. V. 2000. Rous-Whipple award lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am. J. Pathol. 156:1117-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, S. H., K. J. Park, B. Y. Ahn, G. Jung, M. M. Lai, and S. B. Hwang. 2006. Hepatitis C virus nonstructural 5B protein regulates tumor necrosis factor alpha signaling through effects on cellular IκB kinase. Mol. Cell. Biol. 26:3048-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cseh, K., and B. Beutler. 1989. Alternative cleavage of the cachectin/tumor necrosis factor propeptide results in a larger, inactive form of secreted protein. J. Biol. Chem. 264:16256-16260. [PubMed] [Google Scholar]
- 15.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espina, L. M., N. J. Valero, J. M. Hernandez, and J. A. Mosquera. 2003. Increased apoptosis and expression of tumor necrosis factor-alpha caused by infection of cultured human monocytes with dengue virus. Am. J. Trop. Med. Hyg. 68:48-53. [PubMed] [Google Scholar]
- 17.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwarzle, and R. Bartenschlager. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 84:1253-1259. [DOI] [PubMed] [Google Scholar]
- 18.Gilles, P. N., G. Fey, and F. V. Chisari. 1992. Tumor necrosis factor alpha negatively regulates hepatitis B virus gene expression in transgenic mice. J. Virol. 66:3955-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, R. Lew, B. L. Innis, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J. Infect. Dis. 179:755-762. [DOI] [PubMed] [Google Scholar]
- 20.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, A. L. Rothman, and F. A. Ennis. 1999. Elevated plasma interleukin-10 levels in acute dengue correlate with disease severity. J. Med. Virol. 59:329-334. [PubMed] [Google Scholar]
- 21.Gualano, R. C., M. J. Pryor, M. R. Cauchi, P. J. Wright, and A. D. Davidson. 1998. Identification of a major determinant of mouse neurovirulence of dengue virus type 2 using stably cloned genomic-length cDNA. J. Gen. Virol. 79:437-446. [DOI] [PubMed] [Google Scholar]
- 22.Guilhot, S., L. G. Guidotti, and F. V. Chisari. 1993. Interleukin-2 downregulates hepatitis B virus gene expression in transgenic mice by a posttranscriptional mechanism. J. Virol. 67:7444-7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halstead, S. B. 1989. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev. Infect. Dis. 11(Suppl. 4):S830-S839. [DOI] [PubMed] [Google Scholar]
- 24.Halstead, S. B., E. J. O'Rourke, and A. C. Allison. 1977. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J. Exp. Med. 146:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbein, G., and W. A. O'Brien. 2000. Tumor necrosis factor (TNF)-alpha and TNF receptors in viral pathogenesis. Proc. Soc. Exp. Biol. Med. 223:241-257. [DOI] [PubMed] [Google Scholar]
- 26.Ho, L. J., J. J. Wang, M. F. Shaio, C. L. Kao, D. M. Chang, S. W. Han, and J. H. Lai. 2001. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J. Immunol. 166:1499-1506. [DOI] [PubMed] [Google Scholar]
- 27.Hober, D., L. Poli, B. Roblin, P. Gestas, E. Chungue, G. Granic, P. Imbert, J. L. Pecarere, R. Vergez-Pascal, P. Wattre, et al. 1993. Serum levels of tumor necrosis factor-alpha (TNF-alpha), interleukin-6 (IL-6), and interleukin-1 beta (IL-1 beta) in dengue-infected patients. Am. J. Trop. Med. Hyg. 48:324-331. [DOI] [PubMed] [Google Scholar]
- 28.Hober, D., L. Shen, S. Benyoucef, D. De Groote, V. Deubel, and P. Wattre. 1996. Enhanced TNF alpha production by monocytic-like cells exposed to dengue virus antigens. Immunol. Lett. 53:115-120. [DOI] [PubMed] [Google Scholar]
- 29.Jarvis, M. A., J. A. Borton, A. M. Keech, J. Wong, W. J. Britt, B. E. Magun, and J. A. Nelson. 2006. Human cytomegalovirus attenuates interleukin-1β and tumor necrosis factor alpha proinflammatory signaling by inhibition of NF-κB activation. J. Virol. 80:5588-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kittigul, L., W. Temprom, D. Sujirarat, and C. Kittigul. 2000. Determination of tumor necrosis factor-alpha levels in dengue virus infected patients by sensitive biotin-streptavidin enzyme-linked immunosorbent assay. J. Virol. Methods 90:51-57. [DOI] [PubMed] [Google Scholar]
- 31.Kurane, I., U. Kontny, J. Janus, and F. A. Ennis. 1990. Dengue-2 virus infection of human mononuclear cell lines and establishment of persistent infections. Arch. Virol. 110:91-101. [DOI] [PubMed] [Google Scholar]
- 32.Lane, B. R., D. M. Markovitz, N. L. Woodford, R. Rochford, R. M. Strieter, and M. J. Coffey. 1999. TNF-alpha inhibits HIV-1 replication in peripheral blood monocytes and alveolar macrophages by inducing the production of RANTES and decreasing C-C chemokine receptor 5 (CCR5) expression. J. Immunol. 163:3653-3661. [PubMed] [Google Scholar]
- 33.Libraty, D. H., S. Pichyangkul, C. Ajariyakhajorn, T. P. Endy, and F. A. Ennis. 2001. Human dendritic cells are activated by dengue virus infection: enhancement by gamma interferon and implications for disease pathogenesis. J. Virol. 75:3501-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Y. L., C. C. Liu, H. Y. Lei, T. M. Yeh, Y. S. Lin, R. M. Chen, and H. S. Liu. 2000. Infection of five human liver cell lines by dengue-2 virus. J. Med. Virol. 60:425-431. [DOI] [PubMed] [Google Scholar]
- 35.Lin, Y. W., K. J. Wang, H. Y. Lei, Y. S. Lin, T. M. Yeh, H. S. Liu, C. C. Liu, and S. H. Chen. 2002. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 76:12242-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangada, M. M., T. P. Endy, A. Nisalak, S. Chunsuttiwat, D. W. Vaughn, D. H. Libraty, S. Green, F. A. Ennis, and A. L. Rothman. 2002. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J. Infect. Dis. 185:1697-1703. [DOI] [PubMed] [Google Scholar]
- 37.Mangada, M. M., and A. L. Rothman. 2005. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J. Immunol. 175:2676-2683. [DOI] [PubMed] [Google Scholar]
- 38.Marianneau, P., A. Cardona, L. Edelman, V. Deubel, and P. Despres. 1997. Dengue virus replication in human hepatoma cells activates NF-κB which in turn induces apoptotic cell death. J. Virol. 71:3244-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marovich, M., G. Grouard-Vogel, M. Louder, M. Eller, W. Sun, S. J. Wu, R. Putvatana, G. Murphy, B. Tassaneetrithep, T. Burgess, D. Birx, C. Hayes, S. Schlesinger-Frankel, and J. Mascola. 2001. Human dendritic cells as targets of dengue virus infection. J. Investig. Dermatol. Symp. Proc. 6:219-224. [DOI] [PubMed] [Google Scholar]
- 40.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minagawa, H., K. Hashimoto, and Y. Yanagi. 2004. Absence of tumour necrosis factor facilitates primary and recurrent herpes simplex virus-1 infections. J. Gen. Virol. 85:343-347. [DOI] [PubMed] [Google Scholar]
- 42.Mosquera, J. A., J. P. Hernandez, N. Valero, L. M. Espina, and G. J. Anez. 2005. Ultrastructural studies on dengue virus type 2 infection of cultured human monocytes. Virol. J. 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nordeen, S. K. 1988. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques 6:454-458. [PubMed] [Google Scholar]
- 46.Palmer, D. R., P. Sun, C. Celluzzi, J. Bisbing, S. Pang, W. Sun, M. A. Marovich, and T. Burgess. 2005. Differential effects of dengue virus on infected and bystander dendritic cells. J. Virol. 79:2432-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park, K. J., S. H. Choi, S. Y. Lee, S. B. Hwang, and M. M. Lai. 2002. Nonstructural 5A protein of hepatitis C virus modulates tumor necrosis factor alpha-stimulated nuclear factor kappa B activation. J. Biol. Chem. 277:13122-13128. [DOI] [PubMed] [Google Scholar]
- 48.Pavic, I., B. Polic, I. Crnkovic, P. Lucin, S. Jonjic, and U. H. Koszinowski. 1993. Participation of endogenous tumour necrosis factor alpha in host resistance to cytomegalovirus infection. J. Gen. Virol. 74:2215-2223. [DOI] [PubMed] [Google Scholar]
- 49.Peyrefitte, C. N., B. Pastorino, M. Bessaud, H. J. Tolou, and P. Couissinier-Paris. 2003. Evidence for in vitro falsely-primed cDNAs that prevent specific detection of virus negative strand RNAs in dengue-infected cells: improvement by tagged RT-PCR. J. Virol. Methods 113:19-28. [DOI] [PubMed] [Google Scholar]
- 50.Pryor, M. J., J. M. Carr, H. Hocking, A. D. Davidson, P. Li, and P. J. Wright. 2001. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 65:427-434. [DOI] [PubMed] [Google Scholar]
- 51.Raghupathy, R., U. C. Chaturvedi, H. Al-Sayer, E. A. Elbishbishi, R. Agarwal, R. Nagar, S. Kapoor, A. Misra, A. Mathur, H. Nusrat, F. Azizieh, M. A. Khan, and A. S. Mustafa. 1998. Elevated levels of IL-8 in dengue hemorrhagic fever. J. Med. Virol. 56:280-285. [DOI] [PubMed] [Google Scholar]
- 52.Rossol-Voth, R., S. Rossol, K. H. Schutt, S. Corridori, W. de Cian, and D. Falke. 1991. In vivo protective effect of tumour necrosis factor alpha against experimental infection with herpes simplex virus type 1. J. Gen. Virol. 72:143-147. [DOI] [PubMed] [Google Scholar]
- 53.Rothman, A. L., and F. A. Ennis. 1999. Immunopathogenesis of dengue hemorrhagic fever. Virology 257:1-6. [DOI] [PubMed] [Google Scholar]
- 54.Saito, K., K. Meyer, R. Warner, A. Basu, R. B. Ray, and R. Ray. 2006. Hepatitis C virus core protein inhibits tumor necrosis factor alpha-mediated apoptosis by a protective effect involving cellular FLICE inhibitory protein. J. Virol. 80:4372-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott, R. M., A. Nisalak, U. Cheamudon, S. Seridhoranakul, and S. Nimmannitya. 1980. Isolation of dengue viruses from peripheral blood leukocytes of patients with hemorrhagic fever. J. Infect. Dis. 141:1-6. [DOI] [PubMed] [Google Scholar]
- 56.Shresta, S., K. L. Sharar, D. M. Prigozhin, P. R. Beatty, and E. Harris. 2006. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 80:10208-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control, 2nd ed. World Health Organization, Geneva, Switzerland.
- 58.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]