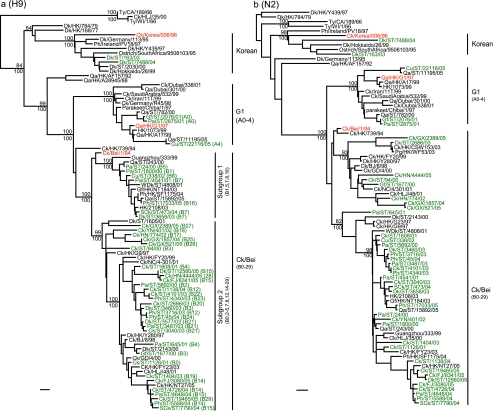
FIG. 3.
Phylogenetic relationships of HA (a) and NA (b) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method in the PAUP* program. Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Analysis was based on nucleotides 129 to 1042 of the HA gene and 231 to 1297 of the NA gene. The HA and NA trees were rooted to Qa/Arkansas/29209-1/93 (H9N2) and Ck/Pennsylvania/8125/83 (H5N2), respectively. Viruses characterized in this study are highlighted in green. Genotypes characterized in this study are shown in parentheses and defined in Table 4. Abbreviations: BJ and Bei, Beijing; Ck, chicken; Dk, duck; FJ, Fujian; GD, Guangdong; Gf, guinea fowl; GX, Guangxi; HK, Hong Kong; HLJ, Heilongjiang; HN, Hunan; NC, Nanchang; Pg, pigeon; Ph, pheasant; Qa, quail; SCk, silky chicken; SD, Shandong; SH, Shanghai; ST, Shantou; Ty, turkey; WDk, wild duck; YN, Yunnan. Bar, 0.01 substitution per site.