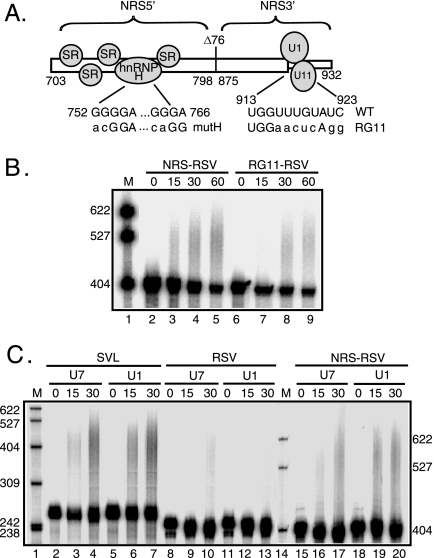
FIG. 3.
U1 and U11 snRNPs are not required for NRS-stimulated RSV polyadenylation. (A) Schematic of the NRS (nt 703 to 932) indicating the NRS5′- (nt 703 to 798), NRS3′- (nt 798 to 932), and NRS-binding factors. Shown are the binding of SR proteins and hnRNP H to NRS5′ and U1 and U11 snRNPs binding to NRS3′. Δ76 indicates a 76-nt deletion that does not markedly affect NRS function. The sequences of mutations that eliminate hnRNP H (mutH) and U1/U11 snRNP (RG11) binding are shown below the wild-type sequence. (B) RSV substrates harboring the wild-type NRS or the RG11 mutation were uniformly labeled with 32P, and polyadenylation was assessed in HeLa cell nuclear extract. (C) SVL, RSV, and NRS-RSV substrates were labeled as described for panel B, and polyadenylation was assayed in HeLa nuclear extract in which U1 or U7 snRNP was inactivated using 2′-O-methyl oligonucleotides. (B and C) Reaction mixtures were incubated for the times (in minutes) indicated above each lane and were subjected to electrophoresis on a 6% polyacrylamide gel that contains 8 M urea. (B) NRS-RSV samples were run ∼1 h longer to allow adequate separation of products. Polyadenylation appears as a slower-migrating smear. Images were obtained with a PhosphorImager and are representative of at least three independent experiments. M, 32P-end-labeled pBR322/MspI markers; WT, wild type. The results of quantitation of polyadenylation at 30 min are the following: (B) NRS-RSV, 9%; and RG11-RSV, 8%; (C) SVL/U7, 45%; SVL/U1, 31%; RSV/U7, 8%; RSV/U1, 5%; NRS RSV/U7, 20%; and NRS RSV/U1, 19%.