Abstract
The geminivirus replication protein AL1 interacts with retinoblastoma-related protein (RBR), a key regulator of the plant division cell cycle, to induce conditions permissive for viral DNA replication. Previous studies of tomato golden mosaic virus (TGMV) AL1 showed that amino acid L148 in the conserved helix 4 motif is critical for RBR binding. In this work, we examined the effect of an L148V mutation on TGMV replication in tobacco cells and during infection of Nicotiana benthamiana plants. The L148V mutant replicated 100 times less efficiently than wild-type TGMV in protoplasts but produced severe symptoms that were delayed compared to those of wild-type infection in plants. Analysis of progeny viruses revealed that the L148V mutation reverted at 100% frequency in planta to methionine, leucine, isoleucine, or a second-site mutation depending on the valine codon in the initial DNA sequence. Similar results were seen with another geminivirus, cabbage leaf curl virus (CaLCuV), carrying an L145A mutation in the equivalent residue. Valine was the predominant amino acid recovered from N. benthamiana plants inoculated with the CaLCuV L145A mutant, while threonine was the major residue in Arabidopsis thaliana plants. Together, these data demonstrated that there is strong selection for reversion of the TGMV L148V and CaLCuV L145A mutations but that the nature of the selected revertants is influenced by both the viral background and host components. These data also suggested that high mutation rates contribute to the rapid evolution of geminivirus genomes in plants.
Geminiviruses constitute the largest and most diverse and economically important family of plant DNA viruses (56). They infect a broad range of plants and cause devastating crop diseases, particularly in tropical and subtropical regions of the world (36, 39, 41). Geminiviruses are characterized by twin icosahedral capsids and small, single-stranded DNA (ssDNA) genomes (56) that display high levels of genetic variability (62). Several studies have indicated that recombination contributes to geminivirus diversity (48, 60, 71). However, unlike double-stranded DNA (dsDNA) viruses, there is also mounting evidence that ssDNA viruses are subject to high nucleotide mutation rates similar to the levels reported previously for RNA viruses (34, 35, 53). Thus, geminiviruses represent a unique opportunity to examine the processes that contribute to the genetic variation of ssDNA viruses as well as the mechanisms underlying virus evolution in plants.
The family Geminiviridae is classified into four genera, Begomovirus, Curtovirus, Topocuvirus, and Mastrevirus, based on their genome organization, host range, and insect vectors (19). The largest genus corresponds to the begomoviruses, which have one- or two-genome components (designated DNA-A and DNA-B), infect dicots, and are transmitted by Bemisia tabaci. Over the past 20 years, there has been a significant increase in the frequency and severity of begomovirus diseases. During this time, agricultural intensification and changes in the insect vector facilitated the expansion of begomovirus populations and their movement into new plant hosts and contributed to the emergence of new, more virulent viruses. Sequence analysis of emerging viruses implicated recombination and reassortment in begomovirus evolution. Both processes depend on the formation of mixed infections and the presence of multiple viral genome components in a single plant cell (54). Recombinant begomoviruses have been associated with severe epidemics in cassava, cotton, and tomato (22, 23, 27, 40, 43, 60, 71) and divergence of the viruses indigenous to the Indian subcontinent (58). Reassortment is a contributing factor to cassava mosaic disease (50), and there are examples of monopartite begomoviruses acquiring DNA-B components (61). In addition, many begomoviruses are associated with DNA satellites that increase virulence and alter host range (7, 37). The satellite DNAs can recombine with themselves and viral genome components (1), further increasing variability.
Nucleotide misincorporation during viral DNA replication also contributes to genome diversity. Studies of bacterial and animal systems indicated that the mutation rates of dsDNA and ssDNA viruses differ significantly. The mutation rates for dsDNA phages range from 10−7 to 10−8, while ssDNA phages display rates of approximately 10−6 (15, 53). Like dsDNA phages, polyomavirus and papillomavirus genomes display low mutation rates (10−8 to 10−9), similarly to their hosts (26). In contrast, high mutation rates (ca. 10−4) have been reported for parvoviruses (35, 64, 65) and circoviruses (6, 21). Like geminiviruses, these viruses have ssDNA genomes that replicate via rolling-circle mechanisms. Thus, the high levels of sequence heterogeneity reported for begomoviruses and mastreviruses (9, 24, 28, 44, 59) may reflect replication errors.
Tomato golden mosaic virus (TGMV) and cabbage leaf curl virus (CaLCuV) are begomoviruses with two-component genomes. Both viruses encode a replication protein designated AL1 (also named AC1, C1, or Rep), which is required for the initiation and termination of viral DNA synthesis (20, 32, 45) and acts as a DNA helicase (11, 12). The AL1 protein also reprograms mature plant cells to create a permissive environment for viral replication through interactions with the host retinoblastoma-related protein (RBR), which regulates cell division and differentiation in plants (14, 17, 31). TGMV and CaLCuV AL1 interact with RBR via a unique 11-amino-acid sequence (2, 31). Alanine substitutions across the helix 4 sequence of TGMV AL1 differentially impacted RBR binding in yeast two-hybrid studies and suggested that residue L148 in the middle of the motif provides a critical binding contact (2, 31). In the experiments reported here, we examined the impact of various amino acid substitutions at TGMV AL1 L148 and the equivalent CaLCuV AL1 L145 on viral replication and infectivity. These studies showed that some mutations reverted at 100% frequency during infection and provided evidence for the capacity of geminivirus populations to evolve rapidly to amend deleterious changes in their genomes.
MATERIALS AND METHODS
AL1 mutants and PCR.
The construction of the TGMV AL1 L148A, L148V, L148M, L148G, and L148I mutations was described previously (2). The mutations are designated by the wild-type residue and its position number followed by the mutant amino acid. TGMV AL1 L148V* and E146A L148V were generated using pNSB148 (46) and primers 5′-TAATTATCTGaAcGGCTTCTTCTTTGGAAGAAGCATTTAAC and 5′-ATTATCTGCAcGGCcgCTTCTTTGGAAGAAGCATTTAA, respectively (lowercase type indicates mutant nucleotides.). TGMV A replicons encoding the mutant AL1 proteins were generated by subcloning SalI/NheI fragments corresponding to AL1 amino acids 120 to 312 from the mutagenesis clones into the same sites of the wild-type replicon pMON1565 (45) to give pNSB919 (E146A/L148V), pNSB979 (L148V), pNSB997 (L148A), pNSB1000 (L148G), and pNSB1031 (L148V*).
TGMV AL1 mutants were also constructed from variants generated during infection with TGMV AL1 L148V, E146A/L148V, or L148V* mutants. Total DNA from systemically infected, symptomatic leaves was amplified by PCR using primers 5′-CGACAAAGACGGAGATACTC and 5′-GTCTCATCTCGTCTGGCACG to give a 281-bp fragment corresponding to TGMV A positions 2006 to 2287. The PCR products were digested with SalI/NcoI and subcloned into the same sites of a modified pBlueScript SK(+) plasmid (Stratagene, Inc.) to generate the intermediate plasmids pNBS1076, pNSB1111, pNSB1077, and pNSB1078. SalI/NheI fragments from these plasmids were then subcloned into the same sites of pMON1565 to generate replicons carrying the AL1 mutations L148I (pNSB1082), L148M (pNSB1113), C128W L148V (pNSB1083), and R125G L148V I155L (pNSB1084).
Wild-type CaLCuV replicons pCpCLCV A.003 and pCpCLCV B.003 contain 1.6 copies of the A and B genomes, respectively (68). The CaLCuV AL1 mutation CaL145A in pNSB1097 (2) was subcloned as an AatII/NsiI fragment (AL1 amino acids 132 to 332) into the equivalent sites of pCpCLCVA.003 to generate the corresponding replicon pNSB1101. Mutant CaLCuV replicons were also generated by PCR of variants produced during infection with the CaLCuV L145A mutant. Total DNA from systemically infected symptomatic leaves was amplified by PCR using primers 5′-GTGAATCCGGGCAGTACAAGGTGTC-3′ and 5′-CCCAGATAAAAACGGAATTCTCTGCC-3 to give an 854-bp fragment between positions 1425 and 2279. The PCR products were digested with AatII/EcoRI and subcloned into the same sites of pCpCLCV A.003 to produce replicons carrying the CaLCuV L145V (pNSB1104), L145A/I167L (pNSB1005), and L145T (pNSB1008) mutations.
Replication and infectivity assays.
Transient replication assays were performed using protoplasts isolated from Nicotiana tabacum (BY2) suspension cells, electroporated, and cultured as described previously (20). Cells were transfected with 5 μg of wild-type or mutant A component DNA from TGMV or CaLCuV and 25 μg of sheared salmon sperm DNA. Total DNA was extracted 72 h after transfection, digested with either DpnI/XhoI (TGMV) or DpnI/EcoRI (CaLCuV), and examined for double- and single-stranded viral DNA accumulation by agarose gel blot analysis using 32P-radiolabeled virus-specific probes against A-component DNA. Double-stranded viral DNA was quantified by phosphorimager analysis. Each replication assay was performed in at least three independent experiments.
Nicotiana benthamiana plants were infected by bombardment or agroinoculation (16, 42), while Arabidopsis thaliana Col-0 rosettes were infected by agroinoculation (66). For bombardment, wild-type or mutant replicon DNA (10 μg) for either TGMV A or CaLCuV A was precipitated onto 1-mm gold microprojectiles in the presence of the corresponding wild-type B-replicon DNA. The wild-type TGMV A and B plasmids were pMON1565 (45) and pTG1.4B (20), while the wild-type CaLCuV A and B plasmids were pCPCBLCVA.003 and pCPCbLCVB.002 (68). For agroinoculation, Agrobacterium tumefaciens cultures carrying a wild-type CaLCuV A (pNSB1090) or a mutant A replicon were mixed with a culture carrying a wild-type CaLCuV B replicon (pNSB1091) and syringe inoculated immediately below the plant apex. Total DNA was extracted from young leaf tissue of individual plants at the indicated times after bombardment (13) and linearized with XhoI (TGMV) or EcoRI (CaLCuV). Total DNA (2.5 μg/lane) was resolved on 1% agarose -Tris-acetate-EDTA gels, transferred onto nylon, and hybridized with a 32P-radiolabeled probe specific for A-component DNA.
Total DNA from infected plants was also amplified by PCR using the primers described above, and the AL1 coding region was sequenced directly. The DNA sequencing chromatograms were examined directly to assess the heterogeneity of the population sequence at individual nucleotide positions.
RESULTS
Virus replication is differentially affected by substitutions at L148.
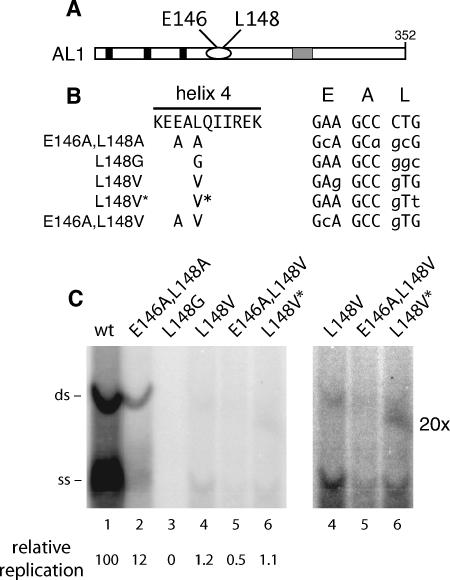
Previously, we showed that valine and glycine substitutions at TGMV AL1 amino acid L148 in the helix 4 motif (Fig. 1A) reduce RBR binding activity to 31 and 36% of wild-type binding in yeast two-hybrid assays (2). To better understand the role of L148 in AL1 function in planta, we examined the impact of valine and glycine mutations on TGMV replication and infectivity assays. We compared the replication of an alanine (E146A L148A), a glycine (L148G), and three valine (L148V, L148V* and E146A L148V) mutants to that of a wild-type replicon (Fig. 1B). L148V and L148V* encode identical proteins but contain one or two nucleotide changes in codon 148, respectively. E146A L148A and E146A L148V are double mutants that also carry alanine substitutions at position E146. E146A L148V has a single nucleotide change at codon 148, like L148V. These mutations were subcloned into the AL1 open reading frame of a replicon plasmid carrying a partial tandem copy of TGMV A with two common regions (55). To ensure maintenance of the mutations, they were subcloned into the unique copy region of the plasmid.
FIG. 1.
L148 mutants are impaired for TGMV AL1 replication. (A) Schematic of the TGMV AL1 protein. Solid boxes mark the locations of the three motifs conserved among rolling-circle replication initiator proteins, the oval indicates a predicted pair of α-helices, and the stippled box shows the location of the ATP binding motif. Helix 4 residues (E146 and L148) that were mutated are indicated. (B) The sequence between TGMV AL1 amino acids 144 and 154 (helix 4) is shown. E146 and L148 substitutions are shown for the five AL1 mutants below the sequence. The codons specifying residues E, A, and L of helix 4 and the mutations introduced into the three codons are shown on the right (modified nucleotides are indicated by lowercase type). Mutants L148V and L148V* differ only in the third position of the codon. (C) Replication of TGMV AL1 mutants was analyzed in tobacco protoplasts by agarose gel blot hybridization. Lanes 1 to 6 are transfections with TGMV A replicons with either wild-type (wt) (lane 1) or mutant AL1 genes corresponding to E146A L148A (lane 2), L148G (lane 3), L148V (lane 4), E146A L148V (lane 5), and L148V* (lane 6). The positions of double-stranded (ds) and single-stranded (ss) forms of TGMV A DNA are marked on the left. An overexposed image (magnification, ×20) of lanes 4 to 6 is shown on the right. The levels of replication of the different mutants relative to wild-type TGMV (100) are indicated at the bottom.
We analyzed the transient replication of wild-type TGMV A and the mutant replicons in N. tabacum BY-2 protoplasts at 72 h posttransfection on agarose gel blots probed with radiolabeled TGMV A DNA. The double-stranded form of the E146A L148A mutant (Fig. 1C, lane 2) accumulated to 12% of wild-type levels (lane 1), similar to the 13% level reported previously for an L148A mutant (2). The L148G mutant (Fig. 1C, lane 3) failed to replicate to detectable levels (Fig. 1C, lane 3). All three valine mutants (Fig. 1C, lanes 4 to 6) were severely impaired for replication, accumulating to ca. 1% of wild-type TGMV A DNA levels. The same TGMV DNA accumulation patterns were observed when tobacco protoplasts were cotransfected with a TGMV B replicon and plant expression cassettes corresponding to the mutant AL1 proteins and wild-type AL3 (data not shown).
The L148 valine mutants develop symptoms and accumulate viral DNA at variable times after inoculation.
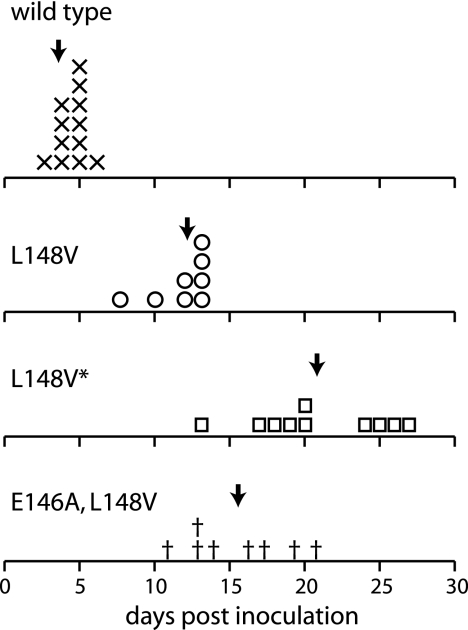
Previous studies showed that N. benthamiana plants infected with the TGMV AL1 helix 4 mutants KEE146 and L148A develop mild chlorosis along the veins 2 to 3 weeks later than plants inoculated with wild-type virus, which showed severe stunting, chlorosis, and leaf curling (2, 31). We asked if the glycine and valine substitutions at position L148 also impact symptoms in infectivity assays. N. benthamiana plants were cobombarded with wild-type TGMV B replicon DNA and either wild-type or mutant A-component DNA, and symptoms were monitored until the plants flowered and set seed at ca. 45 days postinoculation (dpi).
Plants bombarded with wild-type TGMV began to show symptoms at 4 to 5 dpi, with all of the plants displaying severe symptoms by 6 to 7 dpi (Fig. 2). In contrast, none of the plants inoculated with the glycine or valine mutants showed symptoms at 6 dpi in three independent experiments. Consistent with its inability to replicate in tobacco protoplasts, the L148G mutant did not induce any disease symptoms by 45 dpi (data not shown). The three valine mutants were infectious but displayed different kinetics of symptom appearance (Fig. 2). Plants infected with the L148V mutant developed symptoms between 8 and 14 dpi, while plants infected with the E146A L148V mutant began to show symptoms between 9 and 21 dpi. Plants infected with the L148V* mutant showed the greatest delay, with symptoms appearing between 13 and 27 dpi. The average time of symptom appearance was 11.8 dpi for L148V, 15.5 dpi for E146A L148V, and 23.2 dpi for L148V*. In all cases, plants inoculated with the valine mutants eventually developed severe symptoms that were indistinguishable from those induced by wild-type TGMV.
FIG. 2.
Symptom appearance is delayed in plants infected with TGMV L148 valine mutants. N. benthamiana plants cobombarded with either wild-type or mutant TGMV A and wild-type TGMV B replicons were examined daily for the appearance of symptoms in new growth. The dpi when plants displayed unequivocal symptoms (yellow veins and leaf curling) are plotted for each construct. The symbols represent when individual plants displayed symptoms for wild type (×), L148V (○), L148V* (□), and E146A L148V (†). The total number of plants was 12 for the wild type, 8 for L148V, 10 for L148V*, and 9 for E146A L148V. The arrows indicate the average time of symptom appearance for the plants infected with each construct. The data summarize results from two independent experiments.
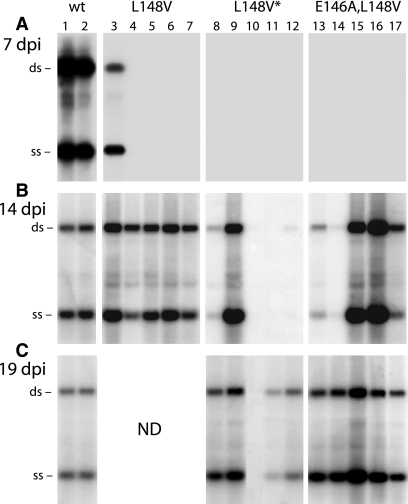
To determine if the time of symptom appearance corresponded to DNA accumulation for the valine mutants, total DNA was isolated from newly emerging leaves of inoculated plants at 7, 14, and 19 dpi and analyzed on agarose gel blots using a radiolabeled TGMV A probe. At 7 dpi, high levels of viral DNA were observed in all plants inoculated with wild-type TGMV (Fig. 3A, lanes 1 and 2), while only one plant infected with the L148V mutant contained detectable levels of TGMV A DNA (lanes 3 to 5), and no plants inoculated with L148V* (lanes 8 to 12) or E146A L148A (lanes 13 to 17) had detectable viral DNA. By 14 dpi, all plants bombarded with the L148V mutant accumulated wild-type levels of viral DNA (Fig. 3B, lanes 3 to 7). At 14 dpi, plants infected with the E146A L148V mutant exhibited variable levels of TGMV DNA in their tissues, ranging from higher-than-wild-type levels (Fig. 3B, lanes 15 and 16) to barely detectable levels (lanes 13 and 14). In contrast, only one of five plants inoculated with the L148V* mutant accumulated substantial amounts of viral DNA at 14 dpi (Fig. 3B, lane 9), although TGMV A DNA was detected at very low levels in two other plants (lanes 8 and 12). At 19 dpi, all plants infected with the E146A L148V mutant (Fig. 3C, lanes 13 to 17) and four of five plants inoculated with the L148V* mutant exhibited high levels of viral DNA in leaves (lanes 8 to 12). In general, plants with detectable TGMV DNA were symptomatic, with the only exception being plants with very low DNA levels (Fig. 3B, lanes 12 and 14). These results were unexpected because of the low replication efficiencies observed for the valine mutants in protoplasts. The results also differed significantly from those reported previously for the KEE146 and L148A mutants, which never accumulated high levels of viral DNA in infected plants over time (2, 31).
FIG. 3.
Viral DNA accumulation is delayed in plants infected with TGMV L148 valine mutants. N. benthamiana plants were bombarded with DNA corresponding to TGMV A and B replicons. The AL1 gene either was the wild type (wt) (lanes 1 and 2) or carried the L148V (lanes 3 to 7), L148V* (lanes 8 to 12), or E146A L148V (lanes 13 to 17) mutation. For each construct, total DNA was isolated from young leaves of the same five plants at 7, 14, and 19 dpi and analyzed by agarose gel blot hybridization. The positions of single-stranded (ss) and double-stranded (ds) forms of TGMV A DNA are marked on the left. ND, not determined.
The L148 valine mutations are unstable in infected plants.
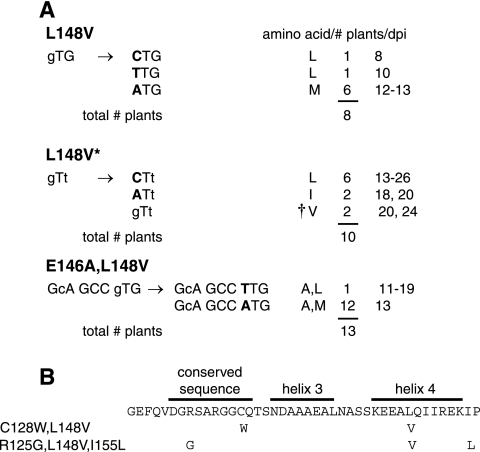
The variability in symptom appearance and viral DNA accumulation and the subsequent development of severe symptoms and high viral DNA levels in plants infected with the L148 valine mutants are consistent with the selection and propagation of a more fit viral variant. We tested this idea by examining the mutated region of the AL1 open reading frame in total DNA extracts from symptomatic young leaves isolated from plants infected with the mutant viruses. A 280-bp fragment encoding TGMV AL1 amino acids 106 to 198, which encompasses the RBR-binding domain (31), was amplified from eight plants infected with the L148V mutant and sequenced directly (Fig. 4A). The L148V codon was modified in all eight plants. In six plants, a G-A transition resulted in a methionine codon at position 148 (Fig. 4A). In the remaining two plants, a G-C or G-T transversion was associated with a reversion of the L148V mutation to leucine.
FIG. 4.
TGMV L148 valine mutants revert at high frequency. Total DNA was isolated from symptomatic leaves of N. benthamiana plants infected with mutant TGMV A and wild-type TGMV B replicons at 19 dpi. The AL1 coding region between amino acids 120 and 180 was amplified from individual plants and sequenced directly. (A) Modifications recovered at codon 148 for L148V, L148V*, and E146A L148V mutants. The original mutations are designated by lowercase type, and the nucleotide changes in the revertants are shown by uppercase, boldface type. The resulting amino acid, the number of plants, and time of symptom appearance (dpi) are shown on the right for each type of revertant. The dagger corresponds to the second-site revertants in B. The total number of plants analyzed for each TGMV mutant is indicated. (B) The TGMV AL1 sequence between amino acids 115 and 156 is shown, with the locations of the predicted α-helices and a conserved sequence marked. Mutations in the second-site revertants are listed on the left, and the amino acid changes are shown below the corresponding positions.
The bias towards methionine substitutions at L148V was also seen in plants inoculated with the E146A L148V mutant (Fig. 4A). In 12 of 13 plants, sequencing uncovered a transition event in which GTG was changed to ATG. Reversion of the L148V mutation to leucine as a consequence of a G-T transversion was seen in only one plant. Interestingly, the E146A mutation in the double mutant was unaltered in all 13 plants (Fig. 4A), indicating that variant selection was highly specific for the L148V codon.
The L148V* codon was also altered at high efficiency during infection, but the sequence changes differed from those detected for the L148V codon (Fig. 4A). The G-A transition found at high frequency in the viral progeny of the L148V and E146A L148V mutants occurred only twice in the 10 L148V* mutant-infected plants. In this case, the GTT codon was altered to ATT to specify an isoleucine. In six plants, a G-C transversion in the first nucleotide of codon 148 took place, resulting in a reversion to leucine (Fig. 4A). Surprisingly, two phenotypic revertants maintained the original L148V* mutation but were altered at other nucleotides in adjacent sequences. One of these second-site revertants displayed a point mutation at codon 128 that substituted a cysteine residue for tryptophan, whereas the other phenotypic revertant with an intact L148V* codon had two changes that generated the substitutions R125G and I155L (Fig. 4B).
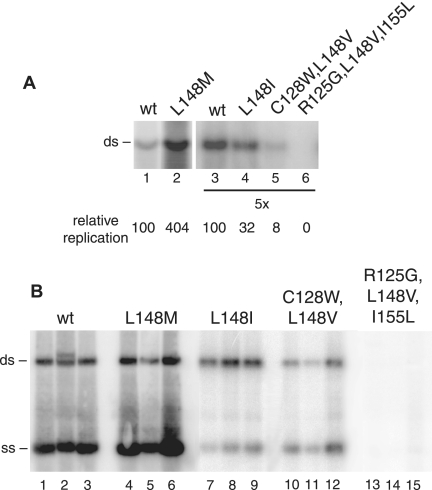
Because only a minority of the valine mutants reverted to the wild-type leucine residue, we asked if the mutations in the sequenced region of the recovered TGMV AL1 mutants were responsible for the observed revertant phenotypes. A fragment encoding TGMV AL1 amino acids 119 to 180 from cloned and sequenced PCR products corresponding to each revertant class was subcloned in place of the homologous region in the wild-type TGMV A replicon. The mutant replicons were tested in transient replication assays (Fig. 5). The L148M mutant (Fig. 5A, lane 2) replicated more efficiently than wild-type TGMV A (lane 1) in tobacco protoplasts, resulting in four times more viral DNA. Viral DNA levels corresponding to the L148I (Fig. 5A, lane 4) and C128W L148V (lane 5) mutations were lower (32% and 8%, respectively) than those of the wild type (lane 3) but significantly higher than those of the original L148V* mutant (Fig. 1C, lane 6). The R125G L148V I155L triple mutant (Fig. 5A, lane 6) did not replicate to readily detected levels in protoplasts, suggesting that other compensatory mutations outside of the subcloned region were responsible for the revertant phenotype.
FIG. 5.
Replication analysis of TGMV revertants. (A) Replication of TGMV A replicons encoding AL1 revertants was analyzed in tobacco protoplasts by agarose gel blot hybridization. Lanes 1 to 6 are transfections with TGMV A replicons with either wild-type (wt) (lanes 1 and 3) or mutant AL1 genes corresponding to L148M (lane 2), L148I (lane 4), C128W L148V (lane 5), and R125G L148V I155L (lane 6). The position of the double-stranded (ds) TGMV A DNA is marked on the left, and levels of replication relative to wild-type TGMV (100) at each exposure are indicated at the bottom of each lane. (B) N. benthamiana plants were cobombarded with wild-type or mutant TGMV A and wild-type TGMV B DNA. At 7 dpi, total DNA was isolated from three individual plants infected with TGMV B and either wild-type TGMV A (lanes 1 to 3) or mutant replicons carrying the L148M (lanes 4 to 6), L148I (lanes 7 to 9), C128W L148V (lanes 10 to 12), or R125G L148V I155L (lanes 13 to 15) mutations. DNA accumulation was monitored by agarose gel blot hybridization. The positions of single-stranded (ss) and double-stranded forms of TGMV A are marked on the left.
The revertant replicons were also assayed for symptom production in N. benthamiana infectivity assays (data not shown). Plants inoculated with the L148M and L148I mutants displayed severe symptoms at 6 dpi. One plant infected with the C128W L148V mutant also showed symptoms at 6 dpi, while three plants displayed symptoms by 8 dpi. Plants inoculated with the R125G L148V I155L mutant developed wild-type symptoms at variable times ranging from 13 to 31 dpi, like the other L148V* mutants (Fig. 2). To verify that the observed symptoms corresponded to viral DNA accumulation, total DNA was isolated from plants at 7 dpi and analyzed on agarose gel blots using a radiolabeled TGMV A probe. High levels of dsDNA and ssDNA were seen in plants inoculated with wild-type TGMV (Fig. 5B, lanes 1 to 3) and the L148M mutant (lanes 4 to 6), while lower amounts of viral DNA were detected for the L148I (lanes 7 to 9) and C128W L148V (lanes 10 to 12) mutants. None of the plants inoculated with the R125G L148V I155L mutant (Fig. 5B, lanes 13 to 15) had detectable amounts of viral DNA at this time point. Together, these results showed that the L148M, L148I, and C128W L148V mutations restored, at least partially, virus replication and infectivity to the valine mutants. The absence of detectable viral DNA early in infection and the development of delayed severe symptoms in combination with the protoplast data suggested that the R125G L148V I155L mutant replicates poorly and undergoes reversion during infection.
The CaLCuV L145A mutant also reverts at high frequency.
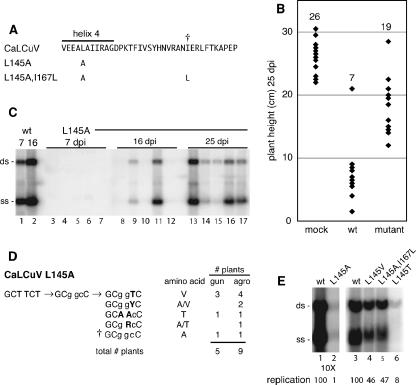
CaLCuV is a begomovirus that is distantly related to TGMV. A previous study showed that the mutation of L145 in the helix 4 motif of the CaLCuV AL1 protein also impairs RBR interactions (2). We asked if an alanine substitution at position L145 negatively impacts CaLCuV infectivity (Fig. 6A), as reported previously for the equivalent TGMV L148A mutation (2). N. benthamiana plants were cobombarded with a wild-type CaLCuV B replicon and either a wild-type CaLCuV A replicon or a mutant A component carrying the L145A substitution. By 4 to 5 dpi, plants inoculated with wild-type virus developed clear symptoms that included leaf curling, vein yellowing, and stunting of new growth (data not shown). The symptoms were more severe than those observed for TGMV-infected plants. In contrast, the five plants inoculated with the CaLCuV L145A mutant did not exhibit any sign of disease at that time. Instead, symptoms appeared in two plants at 15 to 16 dpi, and the remaining plants showed symptoms after 21 dpi. There was a statistically significant reduction in the overall height of CaLCuV-infected plants (Fig. 6B) compared to mock-inoculated plants. Plants inoculated with CaLCuV L145A were less stunted than those infected with wild-type virus but displayed shorter internodes than mock-inoculated plants (Fig. 6B).
FIG. 6.
Reversion of the CaLCuV L145A mutation in N. benthamiana. (A) The sequence of wild-type CaLCuV AL1 from amino acids 141 to 177 is shown. The location of helix 4 and the L145A mutation is indicated. The † symbol marks the amino acid modified in the second-site revertant L145A I167L in D and E. (B) Comparison of the heights of mock-, CaLCuV (wild-type [wt])-, or L145A (mutant)-inoculated N. benthamiana plants at 25 dpi. Each point represents an individual plant, with the mean height for each treatment shown at the top. The means of the three treatments were statistically different (P < 0.01 in a two-tailed Student's t test). (C) N. benthamiana plants were cobombarded with a wild-type CaLCuV B replicon and wild-type CaLCuV A (lanes 1 and 2) or an L145A mutant replicon (lanes 3 to 17). Total DNA from the same five plants at 7, 16, and 25 dpi was analyzed by DNA blot hybridization. The positions of single-stranded (ss) and double-stranded (ds) forms of CaLCuV A DNA are marked on the left. Viral DNA was detected only in plants displaying symptoms. (D) The AL1 coding region between amino acids 132 and 349 was amplified from individual plants and sequenced directly. The original mutation is designated by lowercase type, and the nucleotide changes in the revertants are shown by uppercase, boldface type. Numbers of plants are shown on the right for each type of revertant for bombardment (gun) and agroinoculation (agro) experiments. The total number of plants analyzed for each inoculation protocol is indicated below. (E) Replication of CaLCuV A mutants in tobacco protoplasts was analyzed by agarose gel blot hybridization. Lanes 1 to 6 correspond to transfections with CaLCuV A replicons with either wild-type (lane 1 and 3) or mutant AL1 genes corresponding to L145A (lane 2), L145V (lane 4), L145A I167L (lane 5), and L145T (lane 6). The relative accumulation of viral DNAs is given at the bottom of each lane, with the wild type set at 100 for each exposure.
The delay observed for the CaLCuV L145A mutant is reminiscent of the TGMV L148A mutant, but unlike TGMV L148A, all of the CaLCuV L145A-inoculated plants eventually developed strong symptoms. To better understand this difference, we examined viral DNA accumulation in plants infected with the CaLCuV L145A mutant on agarose gel blots using a radiolabeled CaLCuV A probe (Fig. 6C). At 7 dpi, high levels of viral DNA were detected in plants inoculated with wild-type virus (Fig. 6C, lane 1), while none of the plants infected with the L145A mutant contained detectable levels of viral DNA (lanes 3 to 7). By 16 dpi, three plants infected with the L145A mutant contained detectable levels of viral DNA (Fig. 6C, lanes 8, 9, and 11), and all of the plants were positive for viral DNA by 25 dpi (lanes 13 to 17). The level of viral DNA at 16 and 25 dpi was variable, ranging from high amounts to trace amounts. These results resembled the TGMV DNA accumulation patterns seen for the L148 valine substitutions (Fig. 3), suggesting that the CaLCuV L145A mutation is not stable in plants.
To determine if the L145A mutation is stable, we amplified an 854-bp fragment comprising part of the CaLCuV AL1 and AL2 genes out of total DNA from symptomatic young leaves of five plants infected with the mutant. Sequencing of the PCR products revealed that the original alanine codon was no longer present in four plants (Fig. 6D). In three plants, a C-T transition had occurred to give a valine codon at position 145. In the fourth plant, a GG-AA double transition produced a synonymous mutation at alanine codon 144 and a nonsynonymous mutation at position 145, resulting in a threonine substitution (Fig. 6D). In a fifth plant, the L145A mutation was maintained, but an A-C transversion changed L167 to isoleucine (Fig. 6A and D).
The preferential reversion of CaLCuV L145A to valine was unexpected because a valine substitution at L148 in TGMV AL1 severely impaired replication and infectivity. The CaLCuV and TGMV AL1 proteins show significant divergence in the residues flanking the helix 4 motif, and substitutions at the conserved leucine may differentially impact the replication of the two viruses. To test this possibility, a fragment containing CaLCuV AL1 amino acids 132 to 349 derived from PCR products of each revertant class was subcloned in place of the homologous region in the wild-type CaLCuV A replicon and sequenced prior to transient replication assays (Fig. 6E). The CaLCuV L145V (Fig. 6E, lane 4) and L145A I167L (lane 5) mutants replicated similarly in tobacco protoplasts, accumulating to ca. half of wild-type DNA levels (lane 3). The CaLCuV L145T mutant (Fig. 6E, lane 6) replicated less efficiently, resulting in only 8% of wild-type levels. The L145A (Fig. 6E, lane 6) mutant was the most severely impaired for replication, accumulating at 1% of wild-type levels. This value is significantly less than that reported previously for TGMV L148A, which replicates to 14% of wild-type levels (2), and more similar to that seen for the TGMV L148 valine mutations (Fig. 1C). Consistent with their replication phenotypes, the subcloned CaLCuV L145V, L145A I167L, and L145T replicons were infectious on plants (data not shown).
N. benthamiana plants agroinoculated with wild-type CaLCuV developed severe symptoms indistinguishable from those seen with bombardment (data not shown). The timing of symptom appearance differed between the two protocols, with the agroinoculated plants displaying symptoms 10 to 12 dpi, compared to 4 to 5 dpi for bombarded plants. However, like the bombardment experiments, plants agroinoculated with the CaLCuV L145A mutant were delayed relative to the wild-type control. None of the L145A-inoculated plants exhibited symptoms at 12 dpi, two plants showed signs of disease at 21 dpi, four additional plants displayed symptoms at 23 to 25 dpi, and all nine plants exhibited severe symptoms by 33 dpi.
PCR amplification of viral DNA followed by direct sequencing revealed that the engineered alanine codon was altered in most plants (Fig. 6D). In four plants, the original GCC alanine codon was changed to a GCT valine codon, while a mixture of the two codons was detected in two plants. One plant contained a mixture corresponding to the original GCC codon and a new GTC codon specifying threonine. Another plant retained the original GCC codon but had a second-site L167I mutation, the same second-site reversion found in the bombardment experiment. These results indicated that CaLCuV L145A reversion is not dependent on the inoculation protocol for N. benthamiana.
Different CaLCuV L145A reversions are recovered from Arabidopsis plants.
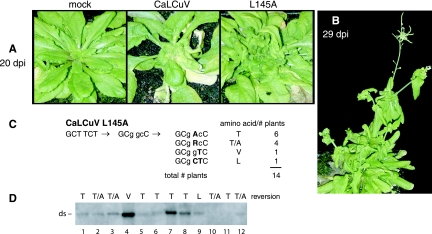
Although CaLCuV infects N. benthamiana, its natural hosts are members of the family Brassicaceae. We took advantage of the ability of CaLCuV to infect Arabidopsis thaliana to ask if the plant host can influence the frequency or the nature of reversion. Arabidopsis plants were agroinoculated with a wild-type CaLCuV B replicon and either a wild-type CaLCuV A replicon or a mutant A component carrying the L145A substitution. At 12 to 15 dpi, all of the plants infected with wild-type CaLCuV developed strong symptoms characterized by yellowing, leaf curling, and severe stunting of new growth. In contrast, none of the 12 plants inoculated with the L145A mutant exhibited symptoms at this time. By 20 dpi, one plant showed mild symptoms, and three additional plants displayed signs of disease by 25 dpi (Fig. 7A). Over the next 8 days, all of the L145A-inoculated plants developed strong symptoms in leaves and flowers (Fig. 7B). These results suggested that the CaLCuV L145A mutation is also unstable in Arabidopsis plants.
FIG. 7.
Reversion of the CaLCuV L145A mutation in Arabidopsis plants. A. thaliana plants agroinoculated with a wild-type CaLCuV B replicon and wild-type CaLCuV A or an L145A mutant replicon are shown. (A) Mock (left), wild-type (middle), and L145A (right) symptoms at 20 dpi. (B) L145A symptoms at 29 dpi. (C) The AL1 coding region between amino acids 132 and 349 was amplified from individual plants and sequenced directly. The original mutations are designated by lowercase type, and the nucleotide changes in the revertants are shown by uppercase, boldface type. The altered amino acid and the numbers of plants are shown on the right for each type of revertant. The total number of plants analyzed is indicated below. (D) Total DNA was isolated from plants at 29 dpi and analyzed by agarose gel blot hybridization. The reversion at L145A is indicated at the top of each lane. The position of double-stranded (ds) CaLCuV A DNA is marked on the left.
An 854-bp viral fragment was amplified from total DNA isolated from symptomatic young leaves of CaLCuV L145A-inoculated Arabidopsis plants at 29 dpi, and the PCR products were sequenced directly. In six plants, a G-A transition at the first nucleotide position changed the engineered alanine codon (GCC) to a threonine codon (ACC) (Fig. 7C). In one plant, a C-T transition at the second nucleotide position resulted in a valine codon (GTC). A double substitution changing GCC to the leucine codon CTC was recovered from a single plant. In four cases, the recovered sequence was a mixture of the engineered GCC codon and the revertant ACC codon, indicative of the presence of two viral variants in the same plant. Together, these results demonstrated a preference for the A145T reversion (10 out 12), with the A145V and A145L reversions occurring at reduced frequency (Fig. 7C). This is in contrast to data for N. benthamiana (Fig. 6D) showing that the A145V reversion occurs more frequently (9/14) than the A145T event (3/14).
The same DNA samples were analyzed by agarose gel blotting using a radiolabeled CaLCuV A-specific probe. A band corresponding to the double-stranded form of CaLCuV A was observed in 10 of the 12 plants (Fig. 7D), while no single-stranded DNA was detected (data not shown). The levels of double-stranded viral DNA varied between plants and did not correlate with revertant type. CaLCuV replication assays in cultured Arabidopsis cells were not successful (data not shown), so it was not possible to distinguish the impact of replication efficiency and the time of reversion on the frequency and accumulation of the revertants. However, the CaLCuV data demonstrated that the plant host influences the outcome of the reversion process but not the overall frequency, which was 100% in all instances.
DISCUSSION
It is generally thought that nucleotide misincorporation does not contribute significantly to the genomic variation of small DNA viruses that are replicated by cellular DNA polymerases (15). This assumption is supported by long-term mutation rates for dsDNA viruses, which are low and comparable to those measured for cellular genes (5). There are numerous reports of the emergence of geminivirus strains with altered pathogenicity (62), indicative of rapid genetic change that has been attributed to recombination or reassortment among different viral genomes. However, our finding that mutations in the helix 4 motif of the AL1 gene of two distantly related begomoviruses revert at 100% frequency suggests that nucleotide substitutions occur with high incidence and are under strong selective pressure during geminivirus infection. Thus, in agreement with recent reports of high mutation rates for other ssDNA viruses infecting vertebrates and bacteria (51, 64, 65), nucleotide substitution events are likely to contribute to the diversity and rapid evolution of geminivirus ssDNA genomes.
Analysis of TGMV and CaLCuV variants with mutations at the equivalent L148 and L145 residues in their respective helix 4 motifs revealed several features of the nucleotide substitution process during geminivirus infection. First, the process is highly efficient, with reversions occurring in 100% of plants infected with either one of the TGMV L148 valine mutants (L148V, L148V*, and E146A L148V) or the CaLCuV L145A mutant. Second, the process is selective for mutations that impair protein function. The E146A mutation, which has no detectable impact on AL1 function (2), was stable even when the valine residue reverted in the E146A L148V mutant. Last, the frequency of a reversion event reflected the number of nucleotide changes required to generate a given amino acid codon. L148V, which has a GTG valine codon, reverted to an ATG methionine codon via a single nucleotide change in 18 of 21 events. The generation of an ATG codon from L148V*, with a GTT valine codon, would have required two changes and was not recovered. Instead, the most common change (6 of 10 events) was to a CTT leucine codon, which also involved a single nucleotide change. Similarly, the low recovery of leucine or methionine revertants (1 of 25) from CaLCuV L145A-infected plants most likely reflected the need for multiple nucleotide changes to generate the requisite codons.
Comparison of TGMV and CaLCuV revertants also uncovered some important differences. The CaLCuV L145A mutant was unstable during infection, while the equivalent L148A mutant of TGMV AL1 was stable (2). Valine was the most frequent reversion recovered from N. benthamiana plants inoculated with the CaLCuV L145A mutant, while TGMV L148V mutants were unstable during infection. These differences cannot be attributed to host effects because N. benthamiana served as the host for both viruses. CaLCuV AL1 is representative of a small group of replication proteins in the SLCV group that lack the conserved element DGRSARGG(C/Q)Q (3). Interestingly, the second-site mutation C125W mapped to this sequence in TGMV AL1. The CaLCuV L145V mutant replicated efficiently in cultured cells (46% of wild-type levels), while a TGMV L148V mutant replicated poorly (1% of wild-type levels). An I167L second-site revertant of CaLCuV L145A, which was recovered twice, also replicated efficiently in culture. Leucine occurs at the equivalent position in TGMV and is the most common residue at this site in other begomovirus replication proteins except for members of the SLCV group, which have branched aliphatic residues (our unpublished observation). However, a TGMV L148A mutant accumulates to only 13% of the wild-type level in cultured cells, while a CaLCuV L145A I167L mutant accumulates to 47% of wild-type levels, suggesting that different sequence constraints on TGMV and CaLCuV AL1 impact virus stability and revertant selection during infection.
The different fates of the CaLCuV L145A mutant in N. benthamiana and Arabidopsis indicated that the plant host also influences the reversion outcome. Like the experiments with N. benthamiana plants, the CaLCuV L145A mutant was unstable in Arabidopsis and reverted at 100% frequency. However, most revertants (10 of 12) contained a threonine substitution at codon 145 in CaLCuV AL1, in contrast to the valine revertants isolated from N. benthamiana plants. The L145T mutant accumulated to 7% of wild-type levels in BY-2 protoplasts, suggesting that the threonine mutant does not replicate efficiently in Nicotiana species, consistent with it being isolated only once from N. benthamiana. The preferential isolation of CaLCuV L145T from Arabidopsis may be indicative of efficient replication in this host. The selection of a valine or a threonine revertant of CaLCuV L145A may reflect different sequence requirements for an AL1/host protein interaction in N. benthamiana versus Arabidopsis. Although no studies have linked the begomovirus AL1 protein to host range (49, 52), the reversion of a tomato yellow leaf curl virus C4 mutant in tomato but not in N. benthamiana has been attributed to different host constraints on systemic movement (29). Mastrevirus replication proteins have also been implicated in host adaptation (33). Interestingly, partial reversion of a three-nucleotide mutation in the RepA coding sequence also occurs at 100% frequency during maize streak virus (MSV) infection (67). Unlike the TGMV and CaLCuV reversions, the MSV reversion was restricted to a single nucleotide transversion event that restored nucleic acid folding but not RepA function.
The 100% reversion frequencies for TGMV, CaLCuV, and MSV mutants and the isolation of second-site revertants imply that the family Geminiviridae is subject to high rates of nucleotide substitution. The high rates may reflect a failure of geminivirus infection to activate the mismatch repair system, which is responsible for the excision and replacement of misincorporated nucleotides during chromosomal replication. Methylated viral DNA is not a good template for replication and transcription (8, 18), and geminiviruses actively interfere with DNA methylation pathways in infected cells (70). As a consequence, viral DNA is undermethylated, making it difficult for the mismatch repair machinery to distinguish between parental and nascent DNA strands. In addition, gene profiling experiments indicated that although other DNA repair pathways are up-regulated during geminivirus infection, the expression of mismatch repair machinery is not increased in CaLCuV-infected Arabidopsis plants (J. T. Ascencio-Ibáñez and L. Hanley-Bowdoin, unpublished result). Together, these results indicated that geminivirus replication products are not corrected by mismatch repair, increasing the likelihood that a mutation will be fixed. Greater genetic variability might facilitate geminivirus adaptation to new hosts and changing environments, ultimately leading to increased viral fitness (57). The failure to recover the TGMV L148V and CaLCuV L145A mutants from most plants suggested that the less fit mutant A component is lost randomly during viral movement, ultimately leading to its disappearance from the population. The 7 of 26 plants carrying mixtures of the CaLCuV L145A mutant and various revertants may reflect intermediates in this process.
TGMV L148 is located in an 80-amino-acid region of AL1 known to mediate oligomerization and binding to AL3, RBR, and other host factors (4, 10, 30, 31, 47, 63). There was no obvious correlation between the effects of the various L148 mutations on RBR binding and viral replication, as illustrated by comparisons of the relative RBR binding (25, 31, and 36%) and replication (13, 1, and 0%) activities of L148A, L148V and L148G, respectively (2). In addition, AL1 oligomerization activity, which is required for viral replication, is only moderately reduced for the L148A, L148V, and L148G mutants (2). Thus, the instability of the valine mutants is not due to reduced oligomerization or RBR binding and instead may reflect the destabilization of the AL1 protein or impaired interactions with a host factor required for viral replication. Strikingly, only leucine, methionine, and isoleucine revertants were recovered, indicating that only a few amino acids are permissible at the L148 position. All three amino acids have large hydrophobic side chains and high probabilities of occurring in α-helices. TGMV AL1 proteins carrying either leucine or methionine display similar functional properties, while the isoleucine variant is moderately reduced in activity. Interestingly, leucine and methionine are the only amino acids found at the equivalent position in the helix 4 motif of all characterized begomovirus and curtovirus replication proteins (data not shown).
An important consequence of high mutation and recombination rates is the continuous production of genetic variation in geminivirus populations. This variability is balanced by a complex set of selection pressures including those associated with intrinsic properties of the virus, such as the maintenance of essential nucleotide structures and replication signals, and selection pressures to maintain crucial interactions with plant hosts and insect vectors. Thus, despite their variation potential, geminiviruses populations exhibit significant genetic stability over time and space, as has been documented for plant RNA viruses that also display high mutation rates (25, 57). Nonetheless, the evolutionary potential of geminiviruses needs to be considered in long-term control strategies, because any disease management effort will result in selective pressure on the virus population to adapt to new circumstances (38, 57). A recent mathematical analysis of the potential impact of disease control strategies concluded that the use of resistant cultivars with reduced within-plant virus titers puts pressure on the target virus to evolve towards a higher multiplication rate (69). The results reported here demonstrated experimentally that geminivirus variants with residual replication capabilities are under strong selective pressure to generate variants that replicate to high titers. Given the large size and genetic heterogeneity of geminivirus populations and their capacity to rapidly change their genomes by recombination and mutation, it will be necessary to devise resistance strategies that prevent virus replication and not simply reduce it because of the risk of generating more harmful variants that overcome resistance.
Acknowledgments
This research was supported by a grant from the National Science Foundation (MCB-0110536 to L.H.-B.) and a postdoctoral fellowship from the PEW Foundation (P0291SC to G.A.-A.).
We thank Marilyn Roossinck (The Samuel Roberts Noble Foundation), Dominique Robertson, Sharon Settlage, and Luisa Lopez-Ochoa (all at NCSU) for their critical comments on the manuscript.
Footnotes
Published ahead of print on 1 August 2007.
REFERENCES
- 1.Amin, I., S. Mansoor, L. Amrao, M. Hussain, S. Irum, Y. Zafar, S. E. Bull, and R. W. Briddon. 2006. Mobilisation into cotton and spread of a recombinant Cotton leaf curl disease satellite. Arch. Virol. 151:2055-2065. [DOI] [PubMed] [Google Scholar]
- 2.Arguello-Astorga, G., L. Lopez-Ochoa, L.-J. Kong, B. M. Orozco, S. B. Settlage, and L. Hanley-Bowdoin. 2004. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma homolog RBR. J. Virol. 78:4817-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arguello-Astorga, G. R., and R. Ruiz-Medrano. 2001. An iteron-related domain is associated to motif 1 in the replication proteins of geminiviruses: identification of potential interacting amino acid-base pairs by a comparative approach. Arch. Virol. 146:1465-1485. [DOI] [PubMed] [Google Scholar]
- 4.Bagewadi, B., S. Chen, S. K. Lal, N. R. Choudhury, and S. K. Mukherjee. 2004. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J. Virol. 78:11890-11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, H. U. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2:140-143. [DOI] [PubMed] [Google Scholar]
- 6.Biagini, P. 2004. Human circoviruses. Vet. Microbiol. 98:95-101. [DOI] [PubMed] [Google Scholar]
- 7.Briddon, R. W., and J. Stanley. 2006. Subviral agents associated with plant single-stranded DNA viruses. Virology 344:198-210. [DOI] [PubMed] [Google Scholar]
- 8.Brough, C. L., W. E. Gardiner, N. M. Inamdar, X. Y. Zhang, M. Ehrlich, and D. M. Bisaro. 1992. DNA methylation inhibits propagation of Tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18:703-712. [DOI] [PubMed] [Google Scholar]
- 9.Bull, S. E., R. W. Briddon, W. S. Sserubombwe, K. Ngugi, P. G. Markham, and J. Stanley. 2006. Genetic diversity and phylogeography of Cassava mosaic viruses in Kenya. J. Gen. Virol. 87:3053-3065. [DOI] [PubMed] [Google Scholar]
- 10.Castillo, A. G., L. J. Kong, L. Hanley-Bowdoin, and E. R. Bejarano. 2004. Interaction between a geminivirus replication protein and the plant sumoylation system. J. Virol. 78:2758-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhury, N. R., P. S. Malik, D. K. Singh, M. N. Islam, K. Kaliappan, and S. K. Mukherjee. 2006. The oligomeric Rep protein of Mungbean yellow mosaic India virus (MYMIV) is a likely replicative helicase. Nucleic Acids Res. 34:6362-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clerot, D., and F. Bernardi. 2006. DNA helicase activity is associated with the replication initiator protein Rep of tomato yellow leaf curl geminivirus. J. Virol. 80:11322-11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dellaporta, S. L., J. Wood, and J. B. Hicks. 1983. A plant DNA minipreparation: version II. Plant Mol. Biol. Rep. 1:19-21. [Google Scholar]
- 14.Desvoyes, B., E. Ramirez-Parra, Q. Xie, N. H. Chua, and C. Gutierrez. 2006. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egelkrout, E., L. Mariconti, R. Cella, D. Robertson, and L. Hanley-Bowdoin. 2002. The activity of the proliferating cell nuclear antigen promoter is differentially regulated by two E2F elements during plant development. Plant Cell 14:3225-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egelkrout, E. M., D. Robertson, and L. Hanley-Bowdoin. 2001. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermak, G., U. Paszkowski, M. Wohlmuth, O. M. Scheid, and J. Paszkowski. 1993. Cytosine methylation inhibits replication of African cassava mosaic virus by two distinct mechanisms. Nucleic Acids Res. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fauquet, C. M., D. M. Bisaro, R. W. Briddon, J. K. Brown, B. D. Harrison, E. P. Rybicki, D. C. Stenger, and J. Stanley. 2003. Revision of taxonomic criteria for species demarcation in the family Geminiviridae, and an updated list of begomovirus species. Arch. Virol. 148:405-421. [DOI] [PubMed] [Google Scholar]
- 20.Fontes, E. P. B., H. J. Gladfelter, R. L. Schaffer, I. T. D. Petty, and L. Hanley-Bowdoin. 1994. Geminivirus replication origins have a modular organization. Plant Cell 6:405-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallian, P., P. Biagini, H. Attoui, J. F. Cantaloube, B. Dussol, Y. Berland, P. de Micco, and X. de Lamballerie. 2002. High genetic diversity revealed by the study of TLMV infection in French hemodialysis patients. J. Med. Virol. 67:630-635. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Andres, S., G. P. Accotto, J. Navas-Castillo, and E. Moriones. 2007. Founder effect, plant host, and recombination shape the emergent population of begomoviruses that cause the Tomato yellow leaf curl disease in the Mediterranean basin. Virology 359:302-312. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Andres, S., F. Monci, J. Navas-Castillo, and E. Moriones. 2006. Begomovirus genetic diversity in the native plant reservoir Solanum nigrum: evidence for the presence of a new virus species of recombinant nature. Virology 350:433-442. [DOI] [PubMed] [Google Scholar]
- 24.Ge, L., J. Zhang, X. Zhou, and L. Hongye. 2007. Population genetic structure and variability of Tomato yellow leaf curl China virus. J. Virol. 81:5902-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbs, A. 1999. Evolution and origins of tobamoviruses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, E. C. 2004. The phylogeography of human viruses. Mol. Ecol. 13:745-756. [DOI] [PubMed] [Google Scholar]
- 27.Idris, A. M., and J. K. Brown. 2002. Molecular analysis of Cotton leaf curl virus-Sudan reveals an evolutionary history of recombination. Virus Genes 24:249-256. [DOI] [PubMed] [Google Scholar]
- 28.Isnard, M., M. Granier, R. Frutos, B. Reynaud, and M. Peterschmitt. 1998. Quasispecies nature of three Maize streak virus isolates obtained through different modes of selection from a population used to assess response to infection of maize cultivars. J. Gen. Virol. 79:3091-3099. [DOI] [PubMed] [Google Scholar]
- 29.Jupin, I., F. Dekouchkovsky, F. Jouanneau, and B. Gronenborn. 1994. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology 204:82-90. [DOI] [PubMed] [Google Scholar]
- 30.Kong, L., and L. Hanley-Bowdoin. 2002. A geminivirus replication protein interacts with a protein kinase and a motor protein that display different expression patterns during plant development and infection. Plant Cell 14:1817-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L., M. S. Pinner, J. W. Davies, and J. Stanley. 1999. Adaptation of the geminivirus bean yellow dwarf virus to dicotyledonous hosts involves both virion-sense and complementary-sense genes. J. Gen. Virol. 80:501-506. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Bueno, A., M. G. Mateu, and J. M. Almendral. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 77:2701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Bueno, A., L. P. Villarreal, and J. M. Almendral. 2006. Parvovirus variation for disease: a difference with RNA viruses? Curr. Top. Microbiol. Immunol. 299:349-370. [DOI] [PubMed] [Google Scholar]
- 36.Mansoor, S., R. W. Briddon, Y. Zafar, and J. Stanley. 2003. Geminivirus disease complexes: an emerging threat. Trends Plant Sci. 8:128-134. [DOI] [PubMed] [Google Scholar]
- 37.Mansoor, S., Y. Zafar, and R. W. Briddon. 2006. Geminivirus disease complexes: the threat is spreading. Trends Plant Sci. 11:209-212. [DOI] [PubMed] [Google Scholar]
- 38.McDonald, B. A., and C. Linde. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40:349-379. [DOI] [PubMed] [Google Scholar]
- 39.Moffat, A. S. 1999. Geminiviruses emerge as serious crop threat. Science 286:1835. [Google Scholar]
- 40.Monci, F., S. Sanchez-Campos, J. Navas-Castillo, and E. Moriones. 2002. A natural recombinant between the geminiviruses Tomato yellow leaf curl Sardinia virus and Tomato yellow leaf curl virus exhibits a novel pathogenic phenotype and is becoming prevalent in Spanish populations. Virology 303:317-326. [DOI] [PubMed] [Google Scholar]
- 41.Morales, F. J., and P. K. Anderson. 2001. The emergence and dissemination of whitefly-transmitted geminiviruses in Latin America—brief review. Arch. Virol. 146:415-441. [DOI] [PubMed] [Google Scholar]
- 42.Nagar, S., T. J. Pedersen, K. Carrick, L. Hanley-Bowdoin, and D. Robertson. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7:705-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ndunguru, J., J. P. Legg, T. A. Aveling, G. Thompson, and C. M. Fauquet. 2005. Molecular biodiversity of cassava begomoviruses in Tanzania: evolution of cassava geminiviruses in Africa and evidence for East Africa being a center of diversity of cassava geminiviruses. Virol. J. 2:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ooi, K., S. Ohshita, I. Ishii, and Y. Tetsukazu. 1997. Molecular phylogeny of geminivirus infecting wild plants in Japan. J. Plant Res. 110:247-257. [Google Scholar]
- 45.Orozco, B. M., and L. Hanley-Bowdoin. 1996. A DNA structure is required for geminivirus origin function. J. Virol. 270:148-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orozco, B. M., and L. Hanley-Bowdoin. 1998. Conserved sequence and structural motifs contribute to the DNA binding and cleavage activities of a geminivirus replication protein. J. Biol. Chem. 273:24448-24456. [DOI] [PubMed] [Google Scholar]
- 47.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 48.Padidam, M., S. Sawyer, and C. M. Fauquet. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218-225. [DOI] [PubMed] [Google Scholar]
- 49.Petty, I. T. D., S. C. Carter, M. R. Morra, J. L. Jeffrey, and H. E. Olivey. 2000. Bipartite geminivirus host adaptation determined cooperatively by coding and noncoding sequences of the genome. Virology 277:429-438. [DOI] [PubMed] [Google Scholar]
- 50.Pita, J. S., V. N. Fondong, A. Sangare, G. W. OtimNape, S. Ogwal, and C. M. Fauquet. 2001. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82:655-665. [DOI] [PubMed] [Google Scholar]
- 51.Poon, A., and L. Chao. 2005. The rate of compensatory mutation in the DNA bacteriophage phiX174. Genetics 170:989-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qin, Y., and I. T. Petty. 2001. Genetic analysis of bipartite geminivirus tissue tropism. Virology 291:311-323. [DOI] [PubMed] [Google Scholar]
- 53.Raney, J. L., R. R. Delongchamp, and C. R. Valentine. 2004. Spontaneous mutant frequency and mutation spectrum for gene A of phiX174 grown in E. coli. Environ. Mol. Mutagen. 44:119-127. [DOI] [PubMed] [Google Scholar]
- 54.Rasheed, M. S., L. A. Selth, A. M. Koltunow, J. W. Randles, and M. A. Rezaian. 2006. Single-stranded DNA of Tomato leaf curl virus accumulates in the cytoplasm of phloem cells. Virology 348:120-132. [DOI] [PubMed] [Google Scholar]
- 55.Rogers, S. G., D. M. Bisaro, R. B. Horsch, R. T. Fraley, N. L. Hoffman, L. Brand, J. S. Elmer, and A. M. Lloyd. 1986. Tomato golden mosaic virus A component DNA replicates autonomously in transgenic plants. Cell 45:593-600. [DOI] [PubMed] [Google Scholar]
- 56.Rojas, M. R., C. Hagen, W. J. Lucas, and R. L. Gilbertson. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361-394. [DOI] [PubMed] [Google Scholar]
- 57.Roossinck, M. J. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 58.Rothenstein, D., D. Haible, I. Dasgupta, N. Dutt, B. L. Patil, and H. Jeske. 2006. Biodiversity and recombination of cassava-infecting begomoviruses from southern India. Arch. Virol. 151:55-69. [DOI] [PubMed] [Google Scholar]
- 59.Sanz, A. I., A. Fraile, J. M. Gallego, J. M. Malpica, and F. Garcia-Arenal. 1999. Genetic variability of natural populations of Cotton leaf curl geminivirus, a single-stranded DNA virus. J. Mol. Evol. 49:672-681. [DOI] [PubMed] [Google Scholar]
- 60.Sanz, A. I., A. Fraile, F. Garcia-Arenal, X. Zhou, D. J. Robinson, S. Khalid, T. Butt, and B. D. Harrison. 2000. Multiple infection, recombination and genome relationships among begomovirus isolates found in cotton and other plants in Pakistan. J. Gen. Virol. 81:1839-1849. [DOI] [PubMed] [Google Scholar]
- 61.Saunders, K., N. Salim, V. R. Mali, V. G. Malathi, R. Briddon, P. G. Markham, and J. Stanley. 2002. Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293:63-74. [DOI] [PubMed] [Google Scholar]
- 62.Seal, S. E., M. J. Jeger, and F. Van den Bosch. 2006. Begomovirus evolution and disease management. Adv. Virus Res. 67:297-316. [DOI] [PubMed] [Google Scholar]
- 63.Settlage, S. B., A. B. Miller, W. Gruissem, and L. Hanley-Bowdoin. 2001. Dual interaction of a geminivirus replication accessory factor with a viral replication protein and a plant cell cycle regulator. Virology 279:570-576. [DOI] [PubMed] [Google Scholar]
- 64.Shackelton, L. A., and E. C. Holmes. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J. Virol. 80:3666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102:379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen, W. H., and L. Hanley-Bowdoin. 2006. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1 and mammalian AMPK activating kinases. Plant Physiol. 142:1642-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shepherd, D. N., D. P. Martin, D. R. McGivern, M. I. Boulton, J. A. Thomson, and E. P. Rybicki. 2005. A three-nucleotide mutation altering the Maize streak virus Rep pRBR-interaction motif reduces symptom severity in maize and partially reverts at high frequency without restoring pRBR-Rep binding. J. Gen. Virol. 86:803-813. [DOI] [PubMed] [Google Scholar]
- 68.Turnage, M. A., N. Muangsan, C. G. Peele, and D. Robertson. 2002. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30:107-114. [DOI] [PubMed] [Google Scholar]
- 69.Van den Bosch, F., G. Akudibilah, S. Seal, and M. Jeger. 2006. Host resistance and the evolutionary response of plant viruses. J. Appl. Ecol. 43:506-516. [Google Scholar]
- 70.Wang, H., K. J. Buckley, X. Yang, R. C. Buchmann, and D. M. Bisaro. 2005. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79:7410-7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, X. P., Y. L. Liu, L. Calvert, C. Munoz, G. W. Otim-Nape, D. J. Robinson, and B. D. Harrison. 1997. Evidence that DNA-A of a geminivirus associated with severe Cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 78:2101-2111. [DOI] [PubMed] [Google Scholar]