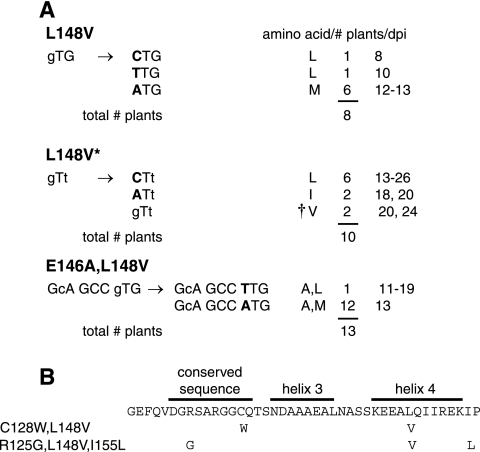
FIG. 4.
TGMV L148 valine mutants revert at high frequency. Total DNA was isolated from symptomatic leaves of N. benthamiana plants infected with mutant TGMV A and wild-type TGMV B replicons at 19 dpi. The AL1 coding region between amino acids 120 and 180 was amplified from individual plants and sequenced directly. (A) Modifications recovered at codon 148 for L148V, L148V*, and E146A L148V mutants. The original mutations are designated by lowercase type, and the nucleotide changes in the revertants are shown by uppercase, boldface type. The resulting amino acid, the number of plants, and time of symptom appearance (dpi) are shown on the right for each type of revertant. The dagger corresponds to the second-site revertants in B. The total number of plants analyzed for each TGMV mutant is indicated. (B) The TGMV AL1 sequence between amino acids 115 and 156 is shown, with the locations of the predicted α-helices and a conserved sequence marked. Mutations in the second-site revertants are listed on the left, and the amino acid changes are shown below the corresponding positions.