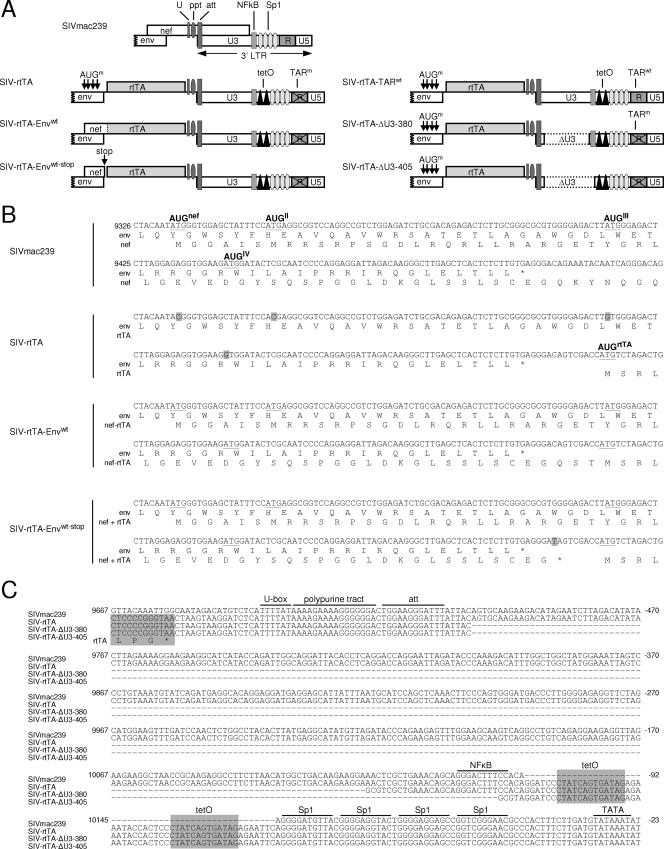
FIG. 4.
Insertion of the rtTA gene and deletion of U3 sequences. In SIVmac239, the 5′ end of the nef open reading frame overlaps the env gene, while the 3′ end overlaps the U-box (U), the PPT, and the U3 region of the LTR promoter (including the attachment sequence required for integration; att). The SIV-rtTA modifications are shown schematically (A) and in detail (B and C). In SIV-rtTA, we replaced the nef sequences downstream of the env gene and upstream of the U-box with the rtTA gene. In addition, we mutated the original AUGNef translation start codon and three downstream AUG codons (AUGII-IV) that precede the new AUGrtTA start codon on the spliced Nef transcript (AUG codons are underlined in panel B). The four AUG mutations (boxed in gray in panel B) were chosen to be synonymous in the env open reading frame and thus not to affect the Env protein. The SIV-rtTA-Envwt variant does not carry these four AUG mutations and will produce a Nef-rtTA fusion protein in which 60 Nef amino acids are fused to the N terminus of the rtTA protein. In the SIV-rtTA-Envwt-stop variant, a translation stop codon was introduced between these Env and rtTA sequences (translation stop codons are indicated with *; mutations are boxed in gray). Accordingly, translation starting at AUGNef will result in the production of a 57-amino-acid Nef polypeptide, while the reinitiation of translation at the AUGrtTA will result in the production of rtTA. In SIV-rtTA-TARwt, the wild-type TAR sequence was reintroduced. In the SIV-rtTA-ΔU3 variants, the Nef-U3 sequences present between the att sequence and either the NF-κB site (SIV-rtTA-ΔU3-380) or the tetO elements (SIV-rtTA-ΔU3-405) were deleted. The numbering is according to the SIVmac239 proviral genome sequence in GenBank/EMBL (accession number M33262; gi 334647).