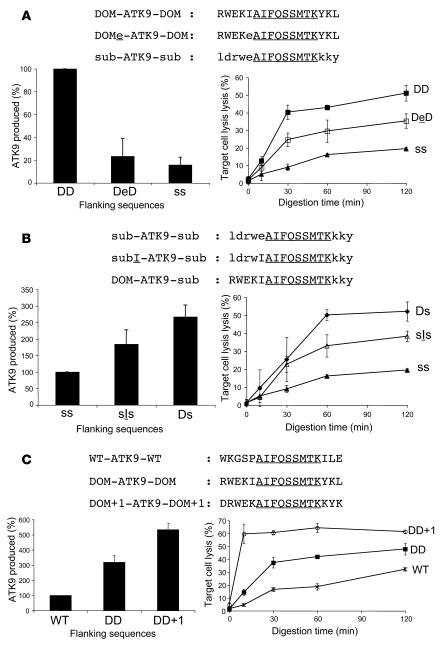
Figure 5. Point mutations in the N terminal portable sequences alter epitope production.
(A) The in vitro degradation of 8 nmol of peptides DOM-ATK9-DOM, DOMe-ATK9-DOM (where flanking I is mutated to E), and sub-ATK9-sub was performed as described in the Figure 4 legend. The amount of ATK9 produced at time 60 was analyzed by mass spectrometry and RP-HPLC. 100% corresponds to the amount of ATK9 produced in 1 hour during DOM-ATK9-DOM degradation (lower left panel). The digestion products from DOM-ATK9-DOM (DD, filled squares), DOMe-ATK9-DOM (DeD, open squares), and sub-ATK9-sub (ss, triangles) were used at 0.05 μg/ml to pulse 51Cr-labeled HLA-A3+ B cells used as targets in a 51Cr release assay using ATK9-specific CTLs. (B) Similar to A except that the 3 peptides were sub-ATK9-sub, subI-ATK9-sub (where flanking E is replaced by I), and DOM-ATK9-sub. 100% corresponds to the amount of ATK9 produced in 1 hour during sub-ATK9-sub degradation. The digestion products from DOM-ATK9-sub (Ds, diamonds), subI-ATK9-sub (sIs, open triangles), and sub-ATK9-sub (ss, triangles) were used in a 51Cr release assay as in A. (C) Similar to A except that the 3 peptides were WT-ATK9-WT, DOM-ATK9-DOM, and DOM+1-ATK9- DOM+1 (Gag flanking sequences shifted by 1 residue). 100% corresponds to the amount of ATK9 produced in 1 hour during WT-ATK9-WT degradation. The digestion products from WT-ATK9-WT (WT, Xs), DOM-ATK9-DOM (DD, squares), and DOM+1-ATK9-DOM+1 (DD+1, circles) were used in a 51Cr release assay as in A. Data are the average of 3 experiments performed with extracts from 3 healthy donors.