Figure 2.

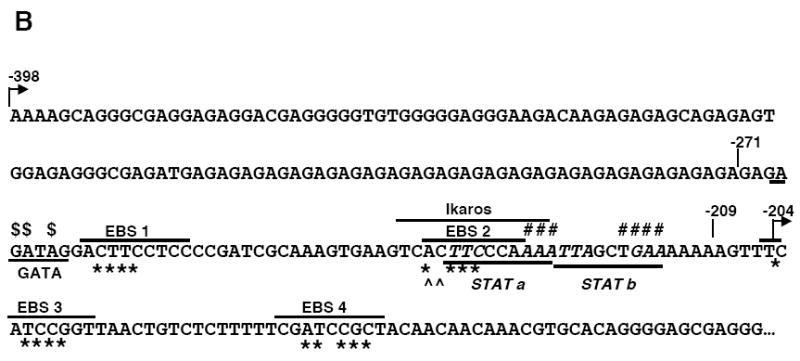
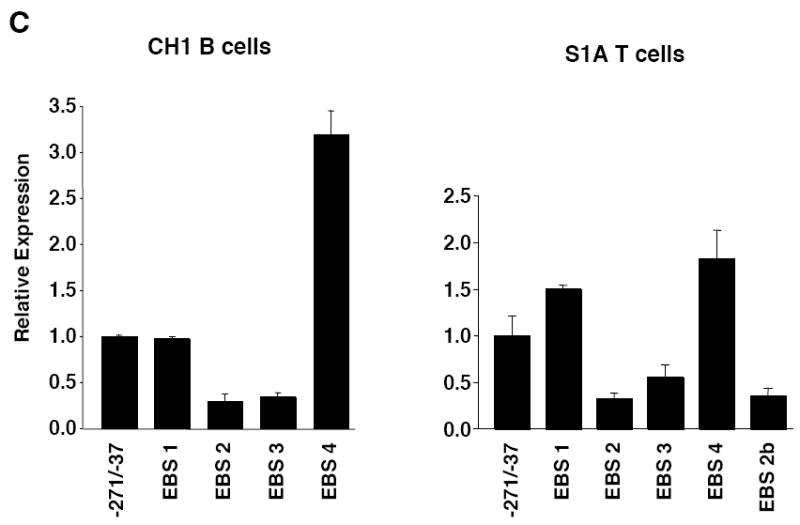
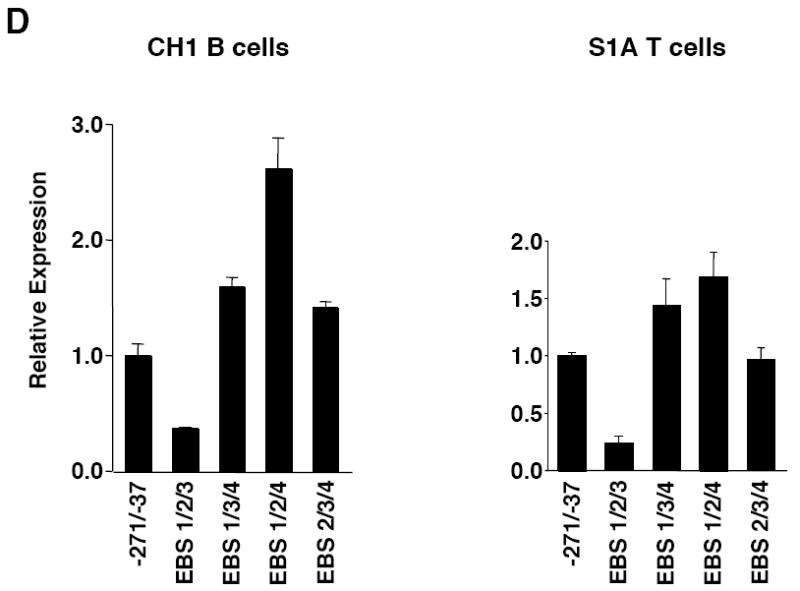
Mutational analysis of four putative Ets binding sites (EBS) in the Fli1 promoter. A) Sequence alignment showing the homology between the murine (M) and human (H) sequence in exon 1 (-398 to +18). Numbering is relative to the +1 translation start site of the murine sequence. Putative transcription factor binding sites are underlined and indicated above the sequence. The coding region of exon 1 is indicated by a shaded box. The mouse sequence was derived from direct sequencing of gDNA cloned from the BALB/c mouse. The human sequence was obtained from NCBI (Y17293.1). B) Location of putative transcription factor binding sites within the -271/-37 region of the murine sequence. Consensus sequences are underlined and mutations made within each consensus site are indicated by: * EBS1, 2, 3 or 4; ^ EBS2b; # STAT; and $ GATA. C) Transient transfection of pGL3 Fli1 -271/-37 containing mutations within each of the four EBSs into CH1 B cells and S1A T cells. D) Transient transfection of pGL3 Fli1 -271/-37 containing combinations of mutations within three of the four EBSs into CH1 B cells and S1A T cells. Results are representative of at least three independent transfections performed with two independently derived clones.
