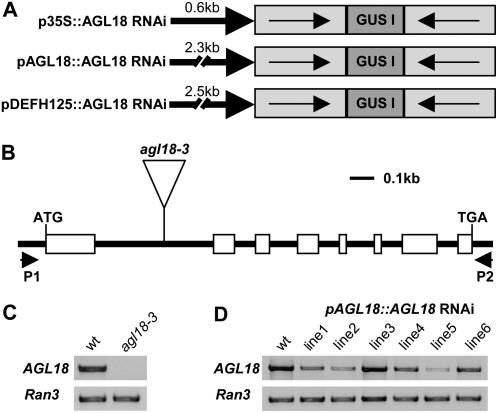
Figure 1.
Gene structure and expression analysis. A, AGL18 RNAi constructs (derivatives of the binary RNAi vector pGSA1252; http://chromdb.org/rnai). AGL18 cDNA (without MADS box, gray boxes) in sense (→) and antisense (←) orientation, separated by a 370-bp GUS intron (GUS I, green box), was cloned into the RNAi cassette driven by three different promoters (bold arrows, promoter length indicated above). B, AGL18 gene structure and T-DNA insertion site of the agl18-3 (NASC ID, N875156) allele. P1 (5′-TAATCTCTTCTCTCTATATCTCTTCTC-3′) and P2 (5′-AGATGAAATAAAGCAAAAGAACAGCCAG-3′) are primers used for reverse transcription-PCR in C and D. C and D, Comparison of AGL18 transcript level between wild-type Columbia plants, agl18-3 T-DNA, and six different RNAi T1 mutants, harboring the AGL18∷AGL18 RNAi construct. Total RNA was isolated from inflorescences of 6-week-old plants grown under long-day conditions (16 h light/8 h dark, 22°C). cDNA was produced from 4 μg of total RNA, treated with RNase-free DNase I (Roche) and using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. PCRs were performed with 31 cycles. Five microliters from 25-μL reactions were loaded on ethidium bromide-stained 1.0% (w/v) agarose gels. Images were scanned with the phosphor imager Typhoon 8600 (Amersham Biosciences). As a control, Ran3 (At5g55190) was amplified using the primers 5′-ACCAGCAAACCGTGGATTACCCTAGC-3′ and 5′-ATTCCACAAAGTGAAGATTAGCGTCC-3′.