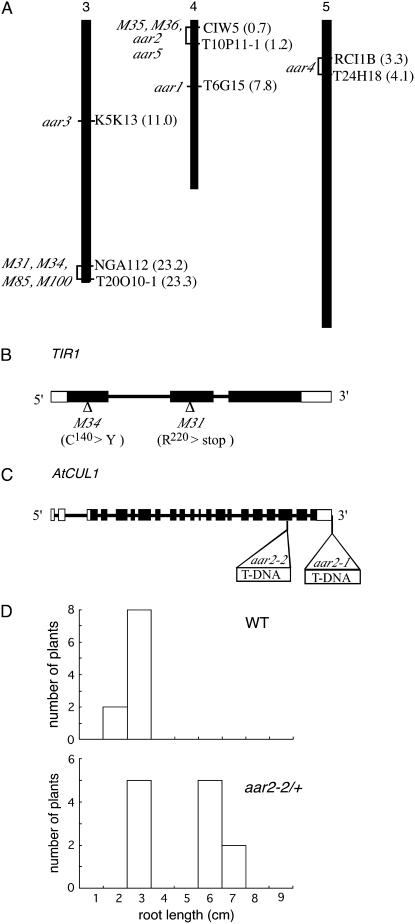
Figure 2.
Mapping of the aar mutants and identification of mutation in TIR1 and AtCUL1 loci. A, Map position of PCIB-resistant mutations. Approximate Arabidopsis Genome Initiative map positions (Mb) of mapping markers are shown in parentheses to the right of each chromosome. The interval to which each PCIB mutant maps is shown to the left of the chromosomes. For aar3-1, no recombinants were identified from 826 chromosomes for markers K5K13-8, K5K13-9, and K5K13-2. m31, m34, m85, and m100 have 1/396, 1/108, 1/20, and 3/104 recombinants at NGA112 and 0/792, 0/110, 0/20, and 0/150 recombinants at T20O10-1, respectively. aar5, m35, and m36 have 6/78, 1/40, and 3/60 recombinants at CIW5-1 and 4/120, 0/86, and 0/60 recombinants at T10P11-1, respectively. aar4 has 1/72 recombinants at both RCI 1B and T24H18 markers. B, Location of the m31 and m34 mutation sites in the TIR1 gene. Black and white boxes indicate exons in translated and untranslated regions, respectively. Lines indicate introns. C, Positions of T-DNA insertions in aar2 mutants in the AtCUL1 gene. D, Distribution of root length of wild-type and aar2-2 heterozygous seedlings germinated and grown vertically on 20 μm PCIB for 14 d.