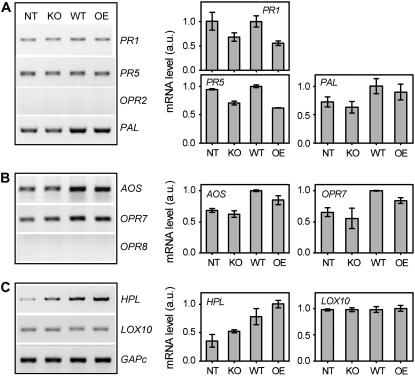
Figure 8.
Expression (sqRT-PCR) of defense-related genes in leaves of T. virens wild type-, deletion strain-, and overexpression strain-induced maize plants challenged with C. graminicola. A, SA-responsive genes: PR1 and PR5, OPR2, and PAL. B, JA-biosynthetic or -inductive genes: AOS, OPR7, and OPR8. C, GLV-biosynthetic or -inductive genes: HPL and LOX10. Expression of GAPc was used as a quantitative standard. The negative control for sqRT-PCR was DNase-treated and cleaned RNA reactions without reverse transcription (not shown). Treatments are: maize seedlings not inoculated with T. virens (NT), or preinoculated with KO25 (KO), Gv29.8 (WT), or OE38 (OE) for 14 d (T = 0, before pathogen challenge). Intensities of bands were quantified using Scion Image software (right panels) and the data expressed as arbitrary units of expression of the indicated defense genes normalized to GAPc. The values shown are the average of two independent experiments with se bars (note: the images of gene expression on the left are from those of the second experiment).