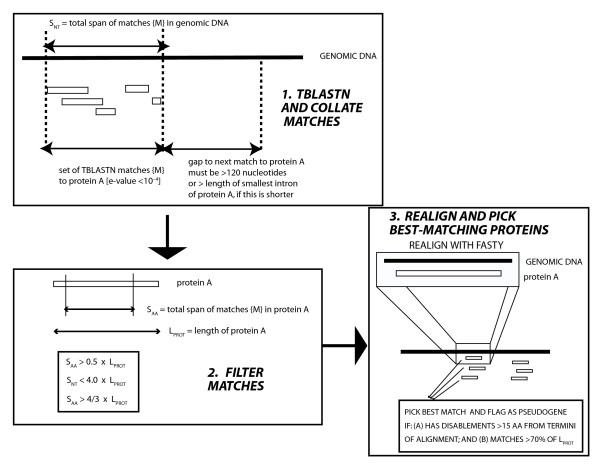
Figure 2.
Rapid annotation of retropseudogenes. (1) TBLASTN matches (e-value ≤ 10-4) of the annotated proteome against the genomic DNA are sorted by coordinates and collated for each protein to form a set of matches {M}. (2) The sets {M} are filtered using length-based heuristics. (3) Each protein is realigned to the genomic DNA using FASTY, and the best-matching proteins at each point have disablements and that matches >70% of the length of the parent sequence are picked as retropseudogene annotations.