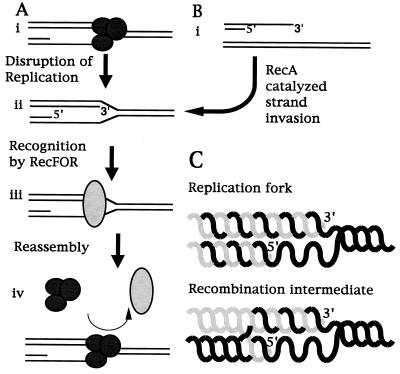
Figure 5.
Model of recF function in vivo. DNA synthesis occurs in a 5′-3′ direction on both strands of duplex DNA. Thus, during semiconservative replication there exists a single-stranded region near the replication fork on the lagging strand template, which will vary in length depending upon where the last lagging strand primer exists (A, i). During genomic replication, if the holoenzyme were to fall off before the completion of replication, the replication fork would be expected to have a structure similar to that shown (ii). Polymerization of the leading strand will terminate with a 3′ end inserted into the homologous double-stranded template DNA. In the simplest model consistent with our results, RecF, RecO, and RecR would recognize this structure as a disrupted replication fork (iii) and facilitate the reassembly of a replication holoenzyme at this structure such that semiconservative DNA synthesis could resume (iv). Such a function for the RecF proteins also could result in recombination when DNA ends are introduced into the system. DNA ends may be present when excessive damage has created strand breaks, when phage DNA has infected the cell, or when DNA has been transfected in artificially. In this situation, recF-dependent recombination is observed to occur when exonucleases process the DNA ends to leave 3′ overhangs (B, i). RecA, which is also required for recF recombination, is known to catalyze the strand invasion of 3′ single-stranded DNA into homologous duplex DNA. If this occurs, the structure created again would be a DNA strand terminating with a 3′ end inserted into homologous duplex DNA as shown (ii). Comparing the resulting structures one finds that they are very similar (C), suggesting that the recF pathway proteins also would recognize this structure. Replication initiated from these DNA ends would incorporate the foreign DNA into the host and result in a recombination event. Such a role for recA in recombination suggests that in vivo it may help maintain the replication fork after holoenzyme disruption.