Abstract
A set of plasmids was constructed so as to contain point mutations which limit the efficiency and/or extent of translation of the gene for ribosomal protein S20. These plasmids were transformed into strains carrying mutations in the genes for polynucleotide phosphorylase (pnp-7), RNase E (rne-1), or both. Subsequently, the effect of translational efficiency on mRNA abundance and chemical half-life was determined. The data indicated that mutations altering translational efficiency also affected mRNA levels over a 10-fold range. This variation in mRNA abundance occurred independently of mutations in either RNase E or polynucleotide phosphorylase, both of which determine the stability of the S20 mRNAs. Moreover, a mutation at codon 15 which caused premature termination of translation of the S20 mRNA did not significantly reduce its stability in different genetic backgrounds. We propose a model in which initiation of translation competes for early steps in mRNA turnover, which could be the binding of RNase E itself or as a complex to one or more sites near the 5' terminus of the S20 mRNA.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet P., Belasco J. G. Control of RNase E-mediated RNA degradation by 5'-terminal base pairing in E. coli. Nature. 1992 Dec 3;360(6403):488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Brosius J., Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis P. P., Nomura M. Regulation of the expression of ribosomal protein genes in Escherichia coli. J Mol Biol. 1975 Sep 5;97(1):61–76. doi: 10.1016/s0022-2836(75)80022-2. [DOI] [PubMed] [Google Scholar]
- Ehretsmann C. P., Carpousis A. J., Krisch H. M. mRNA degradation in procaryotes. FASEB J. 1992 Oct;6(13):3186–3192. doi: 10.1096/fasebj.6.13.1397840. [DOI] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Peltz S. W., Jacobson A. Turnover of mRNA in prokaryotes and lower eukaryotes. Curr Opin Genet Dev. 1992 Oct;2(5):739–747. doi: 10.1016/s0959-437x(05)80134-0. [DOI] [PubMed] [Google Scholar]
- Jain C., Kleckner N. IS10 mRNA stability and steady state levels in Escherichia coli: indirect effects of translation and role of rne function. Mol Microbiol. 1993 Jul;9(2):233–247. doi: 10.1111/j.1365-2958.1993.tb01686.x. [DOI] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. Effects of translation on degradation of mRNA segments transcribed from the polycistronic puf operon of Rhodobacter capsulatus. J Bacteriol. 1991 Feb;173(4):1478–1484. doi: 10.1128/jb.173.4.1478-1484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Cohen S. N. The rate of processing and degradation of antisense RNAI regulates the replication of ColE1-type plasmids in vivo. Cell. 1991 Jun 28;65(7):1233–1242. doi: 10.1016/0092-8674(91)90018-t. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Posttranscriptional regulation of ribosomal protein S20 and stability of the S20 mRNA species. J Bacteriol. 1987 Jun;169(6):2697–2701. doi: 10.1128/jb.169.6.2697-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
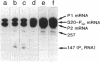
- Mackie G. A. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J Biol Chem. 1992 Jan 15;267(2):1054–1061. [PubMed] [Google Scholar]
- Mackie G. A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J Bacteriol. 1991 Apr;173(8):2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Stabilization of the 3' one-third of Escherichia coli ribosomal protein S20 mRNA in mutants lacking polynucleotide phosphorylase. J Bacteriol. 1989 Aug;171(8):4112–4120. doi: 10.1128/jb.171.8.4112-4120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Structure of the DNA distal to the gene for ribosomal protein S20 in Escherichia coli K12: presence of a strong terminator and an IS1 element. Nucleic Acids Res. 1986 Sep 11;14(17):6965–6981. doi: 10.1093/nar/14.17.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall K. J., Hernandez R. G., Lin-Chao S., Cohen S. N. The ams-1 and rne-3071 temperature-sensitive mutations in the ams gene are in close proximity to each other and cause substitutions within a domain that resembles a product of the Escherichia coli mre locus. J Bacteriol. 1993 Jul;175(13):4245–4249. doi: 10.1128/jb.175.13.4245-4249.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miczak A., Srivastava R. A., Apirion D. Location of the RNA-processing enzymes RNase III, RNase E and RNase P in the Escherichia coli cell. Mol Microbiol. 1991 Jul;5(7):1801–1810. doi: 10.1111/j.1365-2958.1991.tb01929.x. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Carpousis A. J., Krisch H. M. Escherichia coli RNase E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 1990 May;4(5):873–881. doi: 10.1101/gad.4.5.873. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Belasco J. G., Cohen S. N., von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
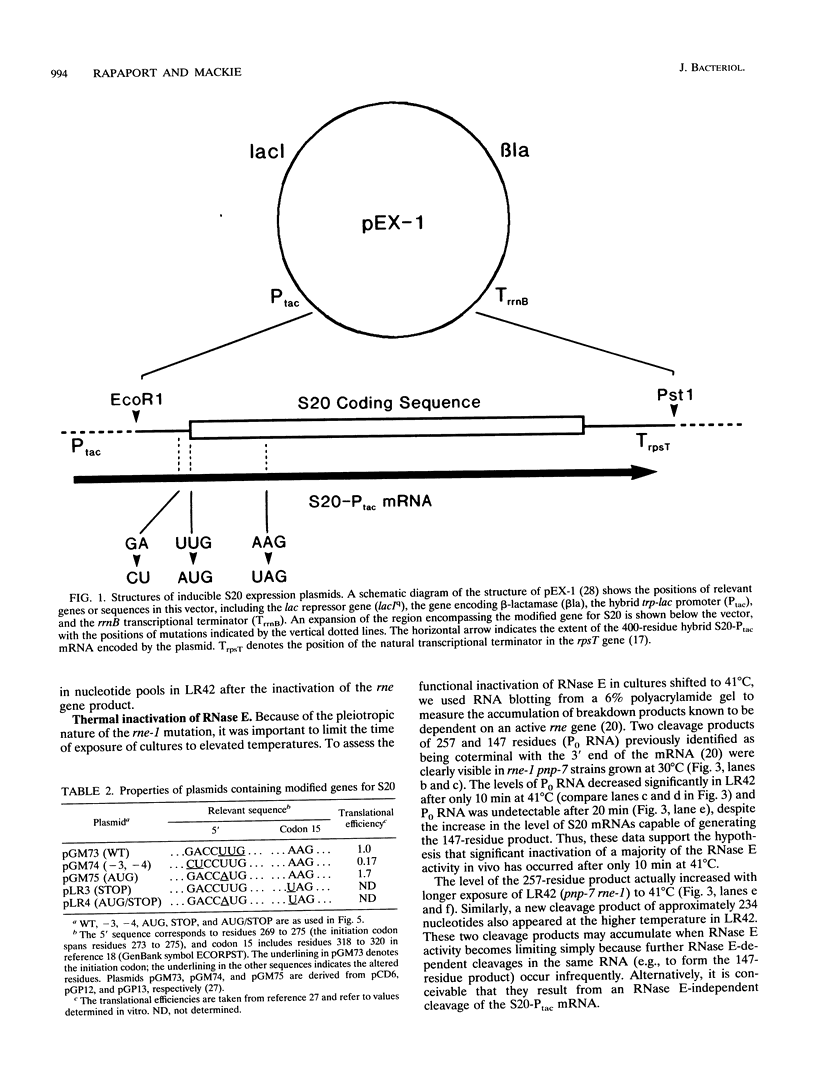
- Parsons G. D., Donly B. C., Mackie G. A. Mutations in the leader sequence and initiation codon of the gene for ribosomal protein S20 (rpsT) affect both translational efficiency and autoregulation. J Bacteriol. 1988 Jun;170(6):2485–2492. doi: 10.1128/jb.170.6.2485-2492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passador L., Linn T. Autogenous regulation of the RNA polymerase beta subunit of Escherichia coli occurs at the translational level in vivo. J Bacteriol. 1989 Nov;171(11):6234–6242. doi: 10.1128/jb.171.11.6234-6242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. Control of functional mRNA stability in bacteria: multiple mechanisms of nucleolytic and non-nucleolytic inactivation. Mol Microbiol. 1992 Feb;6(3):277–282. doi: 10.1111/j.1365-2958.1992.tb01469.x. [DOI] [PubMed] [Google Scholar]
- Yarchuk O., Jacques N., Guillerez J., Dreyfus M. Interdependence of translation, transcription and mRNA degradation in the lacZ gene. J Mol Biol. 1992 Aug 5;226(3):581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]