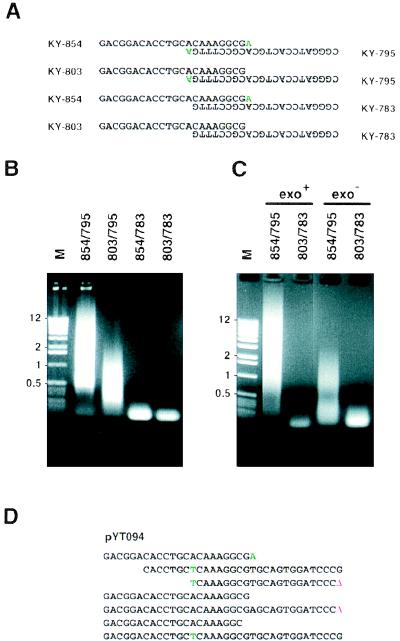
Figure 2.
Effects of the 3′-OH mismatch and 3′–5′ exonuclease activity of DNA polymerase on MPR. (A) Four sets of primers that have double, single, single, and no 3′-OH mismatch nucleotide(s). Mismatch nucleotides are in green. (B) MPR product (10 μl) was fractionated through a 1.2% agarose gel. Lane M, DNA size marker (1-Kb DNA ladder; Life Technologies, Gaithersburg, MD). The sizes of some fragments are indicated on the left (in kilobases). Forty-five cycles of MPR were performed, and the annealing temperature was 69°C. (C) Vent (wt) or Vent (exo−) DNA polymerase was used instead of the Expand Taq polymerase mixture, 45 cycles of MPR were performed, and the annealing temperature was 69°C. (D) An example of a microgene polymer (pYT094) created from KY-854 and KY-795 with Vent (exo−) DNA polymerase. Deletions (Δ) at junctions are in red. Nucleotides originating from the 3′-OH mismatches are in green.