Abstract
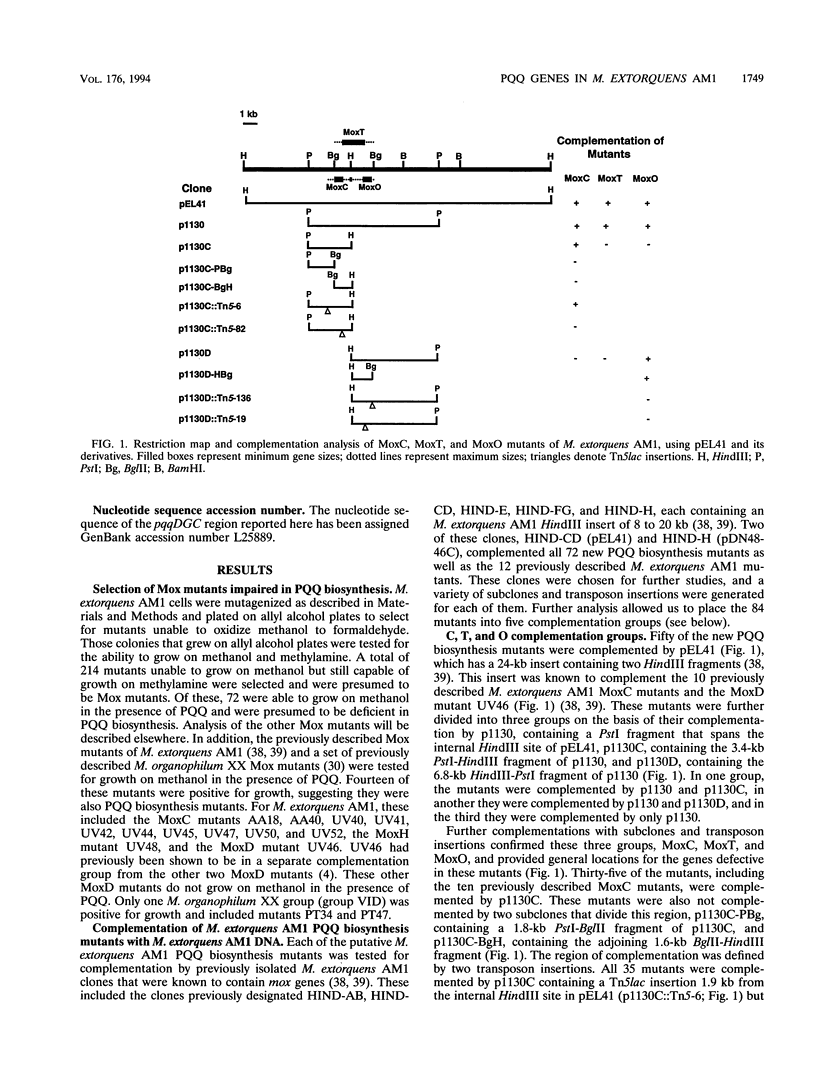
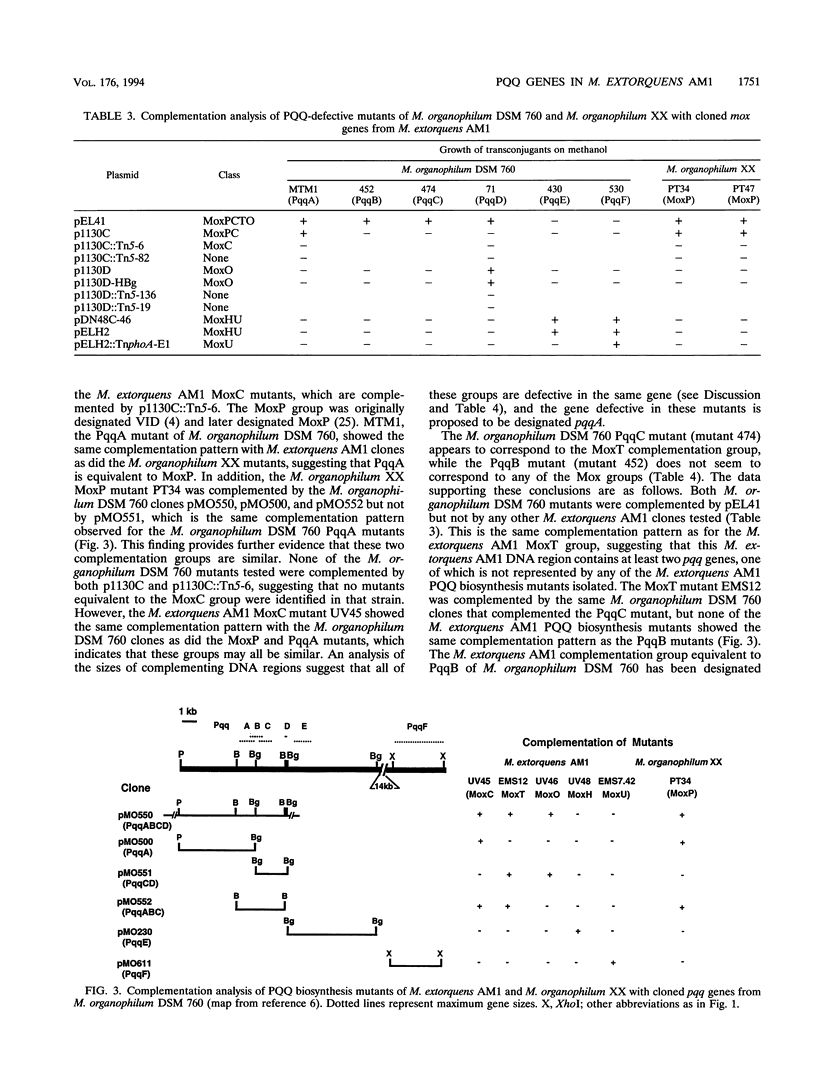
Aerobic gram-negative methylotrophs oxidize methanol to formaldehyde by using a methanol dehydrogenase that has pyrroloquinoline quinone (PQQ) as a prosthetic group. Seventy-two mutants which are unable to grow on methanol unless the growth medium is supplemented with PQQ have been isolated in the facultative methanol utilizer Methylobacterium extorquens AM1. In addition, 12 previously isolated methanol oxidation mutants of M. extorquens AM1 were shown to be able to grow on methanol in the presence of PQQ. These putative PQQ biosynthesis mutants have been complemented by using previously isolated clones containing M. extorquens AM1 DNA, which were known to contain genes necessary for oxidation of methanol to formaldehyde (mox genes). Subcloning and transposon mutagenesis experiments have assigned these mutants to five complementation groups in two gene clusters. Representatives of each complementation group were shown to lack detectable PQQ in the growth medium and in cell extracts and to contain methanol dehydrogenase polypeptides that were inactive. Therefore, these mutants all appear to be defective in PQQ biosynthesis. PQQ biosynthesis mutants of Methylobacterium organophilum DSM 760 and M. organophilum XX were complemented by using M. extorquens AM1 subclones, and PQQ biosynthesis mutants of M. extorquens AM1 and M. organophilum XX were complemented by using M. organophilum DSM 760 subclones. This analysis suggested that a total of six PQQ biosynthesis complementation groups were present in M. extorquens AM1 and M. organophilum DSM 760. A 2-kb M. extorquens AM1 DNA fragment that complemented the MoxO class of PQQ biosynthesis mutants was sequenced and found to contain two complete open reading frames and the N-terminal sequence of a third. These genes designated pqqDGC, had predicted gene products with substantial similarity to the gene products of corresponding pqq genes in Acinetobacter calcoaceticus and Klebsiella pneumoniae. pqqD encodes a 29-amino-acid peptide which contains a tyrosine residue and glutamate residue that are conserved in the equivalent peptides of K. pneumoniae, PqqA (23 amino acids), and A. calcoaceticus, PqqIV (24 amino acids), and are thought to be the precursors for PQQ biosynthesis. The organizations of a cluster of five PQQ biosynthetic genes appear to be similiar in four different bacteria (M. extorquens AM1, M. organophilum DSM 760, K. pneumoniae, and A. calcoaceticus). Our results show that a total of seven pqq genes are present in M. extorquens AM1, and these have been designated pqqDGCBA and pqqEF.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen L. N., Hanson R. S. Construction of broad-host-range cosmid cloning vectors: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985 Mar;161(3):955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. J., Lidstrom M. E. The moxFG region encodes four polypeptides in the methanol-oxidizing bacterium Methylobacterium sp. strain AM1. J Bacteriol. 1988 May;170(5):2254–2262. doi: 10.1128/jb.170.5.2254-2262.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C., Zatman L. J. The microbial oxidation of methanol. 1. Isolation and properties of Pseudomonas sp. M27. Biochem J. 1964 Sep;92(3):609–614. doi: 10.1042/bj0920609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien C., Machlin S., Zhang Y., Donaldson K., Hanson R. S. Organization of Genes Required for the Oxidation of Methanol to Formaldehyde in Three Type II Methylotrophs. Appl Environ Microbiol. 1989 Dec;55(12):3124–3130. doi: 10.1128/aem.55.12.3124-3130.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biville F., Turlin E., Gasser F. Mutants of Escherichia coli producing pyrroloquinoline quinone. J Gen Microbiol. 1991 Aug;137(8):1775–1782. doi: 10.1099/00221287-137-8-1775. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goosen N., Horsman H. P., Huinen R. G., van de Putte P. Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrrolo-quinoline-quinone: nucleotide sequence and expression in Escherichia coli K-12. J Bacteriol. 1989 Jan;171(1):447–455. doi: 10.1128/jb.171.1.447-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen B. W., van Kleef M. A., Duine J. A. Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J. 1986 Mar 15;234(3):611–615. doi: 10.1042/bj2340611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms N., Reijnders W. N., Anazawa H., van der Palen C. J., van Spanning R. J., Oltmann L. F., Stouthamer A. H. Identification of a two-component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993 May;8(3):457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- Heffron F., Bedinger P., Champoux J. J., Falkow S. Deletions affecting the transposition of an antibiotic resistance gene. Proc Natl Acad Sci U S A. 1977 Feb;74(2):702–706. doi: 10.1073/pnas.74.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988 May;119(1):9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lidstrom M. E. Genetics of carbon metabolism in methylotrophic bacteria. FEMS Microbiol Rev. 1990 Dec;7(3-4):431–436. doi: 10.1111/j.1574-6968.1990.tb04949.x. [DOI] [PubMed] [Google Scholar]
- Lidstrom M. E., Stirling D. I. Methylotrophs: genetics and commercial applications. Annu Rev Microbiol. 1990;44:27–58. doi: 10.1146/annurev.mi.44.100190.000331. [DOI] [PubMed] [Google Scholar]
- Machlin S. M., Tam P. E., Bastien C. A., Hanson R. S. Genetic and physical analyses of Methylobacterium organophilum XX genes encoding methanol oxidation. J Bacteriol. 1988 Jan;170(1):141–148. doi: 10.1128/jb.170.1.141-148.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclennan D. G., Ousby J. C., Vasey R. B., Cotton N. T. The influence of dissolved oxygen on Pseudomonas AM1 grown on methanol in continuous culture. J Gen Microbiol. 1971 Dec;69(3):395–404. doi: 10.1099/00221287-69-3-395. [DOI] [PubMed] [Google Scholar]
- Mazodier P., Biville F., Turlin E., Gasser F. Localization of a pyrroloquinoline quinone biosynthesis gene near the methanol dehydrogenase structural gene in Methylobacterium organophilum DSM 760. J Gen Microbiol. 1988 Sep;134(9):2513–2524. doi: 10.1099/00221287-134-9-2513. [DOI] [PubMed] [Google Scholar]
- Meulenberg J. J., Sellink E., Riegman N. H., Postma P. W. Nucleotide sequence and structure of the Klebsiella pneumoniae pqq operon. Mol Gen Genet. 1992 Mar;232(2):284–294. doi: 10.1007/BF00280008. [DOI] [PubMed] [Google Scholar]
- Morris C. J., Lidstrom M. E. Cloning of a methanol-inducible moxF promoter and its analysis in moxB mutants of Methylobacterium extorquens AM1rif. J Bacteriol. 1992 Jul;174(13):4444–4449. doi: 10.1128/jb.174.13.4444-4449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Day D., Anthony C. The second subunit of methanol dehydrogenase of Methylobacterium extorquens AM1. Biochem J. 1989 Jun 15;260(3):857–862. doi: 10.1042/bj2600857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):591–597. doi: 10.1128/jb.166.2.591-597.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson I. W., Anthony C. Characterization of mutant forms of the quinoprotein methanol dehydrogenase lacking an essential calcium ion. Biochem J. 1992 Nov 1;287(Pt 3):709–715. doi: 10.1042/bj2870709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlin E., Biville F., Gasser F. Complementation of Methylobacterium organophilum mutants affected in pyrroloquinoline quinone biosynthesis genes pqqE and pqqF by cloned Escherichia coli chromosomal DNA. FEMS Microbiol Lett. 1991 Sep 15;67(1):59–63. doi: 10.1016/0378-1097(91)90444-f. [DOI] [PubMed] [Google Scholar]
- Van Spanning R. J., Wansell C. W., De Boer T., Hazelaar M. J., Anazawa H., Harms N., Oltmann L. F., Stouthamer A. H. Isolation and characterization of the moxJ, moxG, moxI, and moxR genes of Paracoccus denitrificans: inactivation of moxJ, moxG, and moxR and the resultant effect on methylotrophic growth. J Bacteriol. 1991 Nov;173(21):6948–6961. doi: 10.1128/jb.173.21.6948-6961.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. H., Viebahn M., Hanson R. S. Identification of methanol-regulated promoter sequences from the facultative methylotrophic bacterium Methylobacterium organophilum XX. J Gen Microbiol. 1993 Apr;139(4):743–752. doi: 10.1099/00221287-139-4-743. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



