Abstract
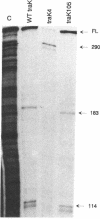
The sequence of traK gene of the F sex factor of Escherichia coli is presented; the traK gene product is predicted to be a protein of 25,627 Da with a signal sequence of 21 amino acids to give a mature protein of 23,307 Da. The traK4 mutation is an extremely polar mutation in the F plasmid that affects F pilus synthesis and plasmid transfer. traK genes carrying the traK4 mutation and a nonpolar mutation traK105 were cloned, sequenced, and identified as an amber nonsense and a frameshift mutation, respectively. The traK4 mutation occurred within one predicted rho-dependent transcription termination element (TTE) and immediately upstream of another, while the traK105 mutation occurred after the two potential TTEs within the traK gene. S1 nuclease protection analysis and Northern (RNA) blot analysis were used to confirm that the traK4 mutation, but not the traK105 mutation, caused premature termination of transcription. Computer analysis of the F transfer region suggested the presence of TTE motifs at regular intervals throughout the 33.4-kb sequence.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Manning P. A., Edelbluth C., Herrlich P. Export without proteolytic processing of inner and outer membrane proteins encoded by F sex factor tra cistrons in Escherichia coli minicells. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4837–4841. doi: 10.1073/pnas.76.10.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Conjugational complementation analysis of transfer-deficient mutants of Flac in Escherichia coli. J Bacteriol. 1972 Jun;110(3):831–842. doi: 10.1128/jb.110.3.831-842.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano P., Ciampi M. S., Nappo A. G., Bruni C. B., Carlomagno M. S. In vivo analysis of the mechanisms responsible for strong transcriptional polarity in a "sense" mutant within an intercistronic region. Cell. 1988 Oct 21;55(2):351–360. doi: 10.1016/0092-8674(88)90058-x. [DOI] [PubMed] [Google Scholar]
- Alifano P., Rivellini F., Limauro D., Bruni C. B., Carlomagno M. S. A consensus motif common to all Rho-dependent prokaryotic transcription terminators. Cell. 1991 Feb 8;64(3):553–563. doi: 10.1016/0092-8674(91)90239-u. [DOI] [PubMed] [Google Scholar]
- Ciampi M. S., Alifano P., Nappo A. G., Bruni C. B., Carlomagno M. S. Features of the rho-dependent transcription termination polar element within the hisG cistron of Salmonella typhimurium. J Bacteriol. 1989 Aug;171(8):4472–4478. doi: 10.1128/jb.171.8.4472-4478.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Paranchych W., Willetts N. S. DNA sequence of the F traALE region that includes the gene for F pilin. J Bacteriol. 1984 Oct;160(1):395–401. doi: 10.1128/jb.160.1.395-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L., Lee S., Yanchar N., Paranchych W. finP and fisO mutations in FinP anti-sense RNA suggest a model for FinOP action in the repression of bacterial conjugation by the Flac plasmid JCFL0. Mol Gen Genet. 1989 Jul;218(1):152–160. doi: 10.1007/BF00330578. [DOI] [PubMed] [Google Scholar]
- Ham L. M., Cram D., Skurray R. Transcriptional analysis of the F plasmid surface exclusion region: mapping of traS, traT, and traD transcripts. Plasmid. 1989 Jan;21(1):1–8. doi: 10.1016/0147-619x(89)90081-4. [DOI] [PubMed] [Google Scholar]
- Ippen-Ihler K., Achtman M., Willetts N. Deletion map of the Escherichia coli K-12 sex factor F: the order of eleven transfer cistrons. J Bacteriol. 1972 Jun;110(3):857–863. doi: 10.1128/jb.110.3.857-863.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen-Ihler K., Moore D., Laine S., Johnson D. A., Willetts N. S. Synthesis of F-pilin polypeptide in the absence of F traJ product. Plasmid. 1984 Mar;11(2):116–129. doi: 10.1016/0147-619x(84)90017-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991 Mar 22;64(6):1047–1049. doi: 10.1016/0092-8674(91)90257-y. [DOI] [PubMed] [Google Scholar]
- Richardson J. P. Rho-dependent transcription termination. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):127–138. doi: 10.1016/0167-4781(90)90048-7. [DOI] [PubMed] [Google Scholar]
- Rivellini F., Alifano P., Piscitelli C., Blasi V., Bruni C. B., Carlomagno M. S. A cytosine- over guanosine-rich sequence in RNA activates rho-dependent transcription termination. Mol Microbiol. 1991 Dec;5(12):3049–3054. doi: 10.1111/j.1365-2958.1991.tb01864.x. [DOI] [PubMed] [Google Scholar]
- Ruteshouser E. C., Richardson J. P. Identification and characterization of transcription termination sites in the Escherichia coli lacZ gene. J Mol Biol. 1989 Jul 5;208(1):23–43. doi: 10.1016/0022-2836(89)90085-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skurray R. A., Nagaishi H., Clark A. J. Construction and BamHL analysis of chimeric plasmids containing EcoRL DNA fragments of the F sex factor. Plasmid. 1978 Feb;1(2):174–186. doi: 10.1016/0147-619x(78)90037-9. [DOI] [PubMed] [Google Scholar]
- Stark M. J. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51(2-3):255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R., Achtman M. The control region of the F sex factor DNA transfer cistrons: physical mapping by deletion analysis. Mol Gen Genet. 1979 Jan 16;169(1):49–57. doi: 10.1007/BF00267544. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Sameshima J. H., Hatfield G. W. Rho-dependent transcriptional polarity in the ilvGMEDA operon of wild-type Escherichia coli K12. J Biol Chem. 1987 Nov 5;262(31):15256–15261. [PubMed] [Google Scholar]
- Willetts N., Achtman M. Genetic analysis of transfer by the Escherichia coli sex factor F, using P1 transductional complementation. J Bacteriol. 1972 Jun;110(3):843–851. doi: 10.1128/jb.110.3.843-851.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N., Maule J., McIntire S. The genetic locations of traO, finP and tra-4 on the E. coli K12 sex factor F. Genet Res. 1975 Dec;26(3):255–263. doi: 10.1017/s0016672300016050. [DOI] [PubMed] [Google Scholar]
- Willetts N. The transcriptional control of fertility in F-like plasmids. J Mol Biol. 1977 May 5;112(1):141–148. doi: 10.1016/s0022-2836(77)80161-7. [DOI] [PubMed] [Google Scholar]
- Yager T. D., von Hippel P. H. A thermodynamic analysis of RNA transcript elongation and termination in Escherichia coli. Biochemistry. 1991 Jan 29;30(4):1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]