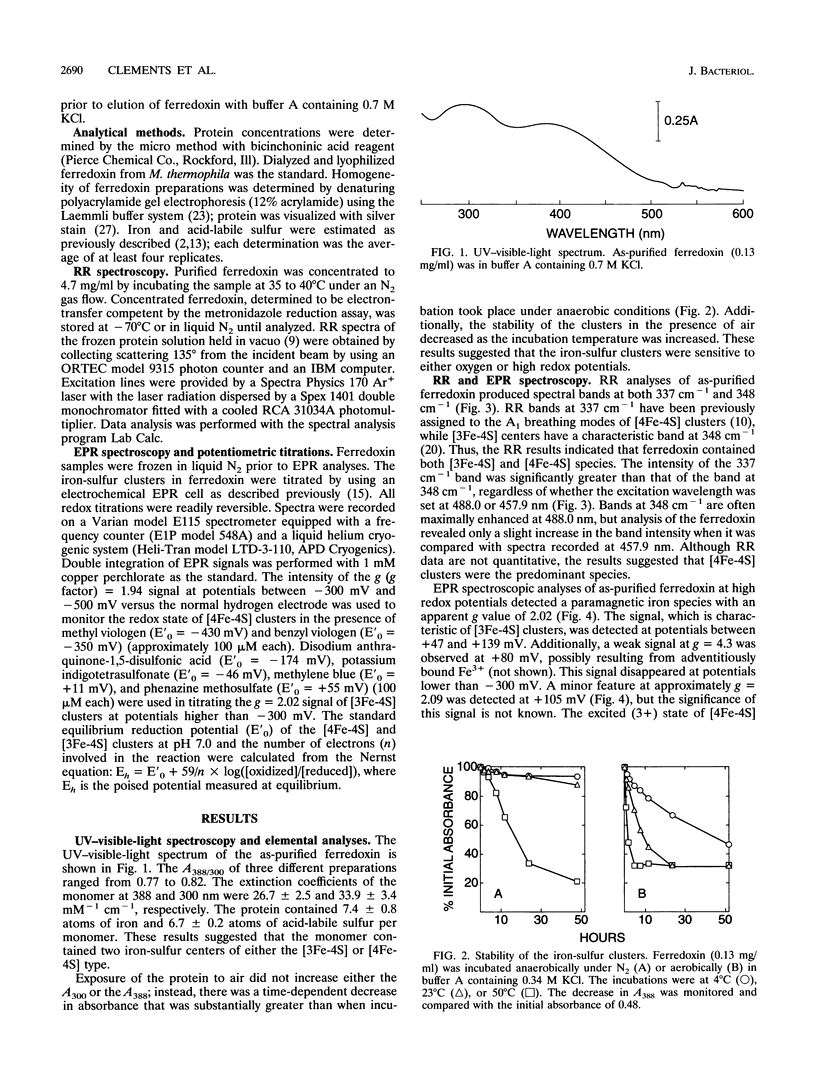
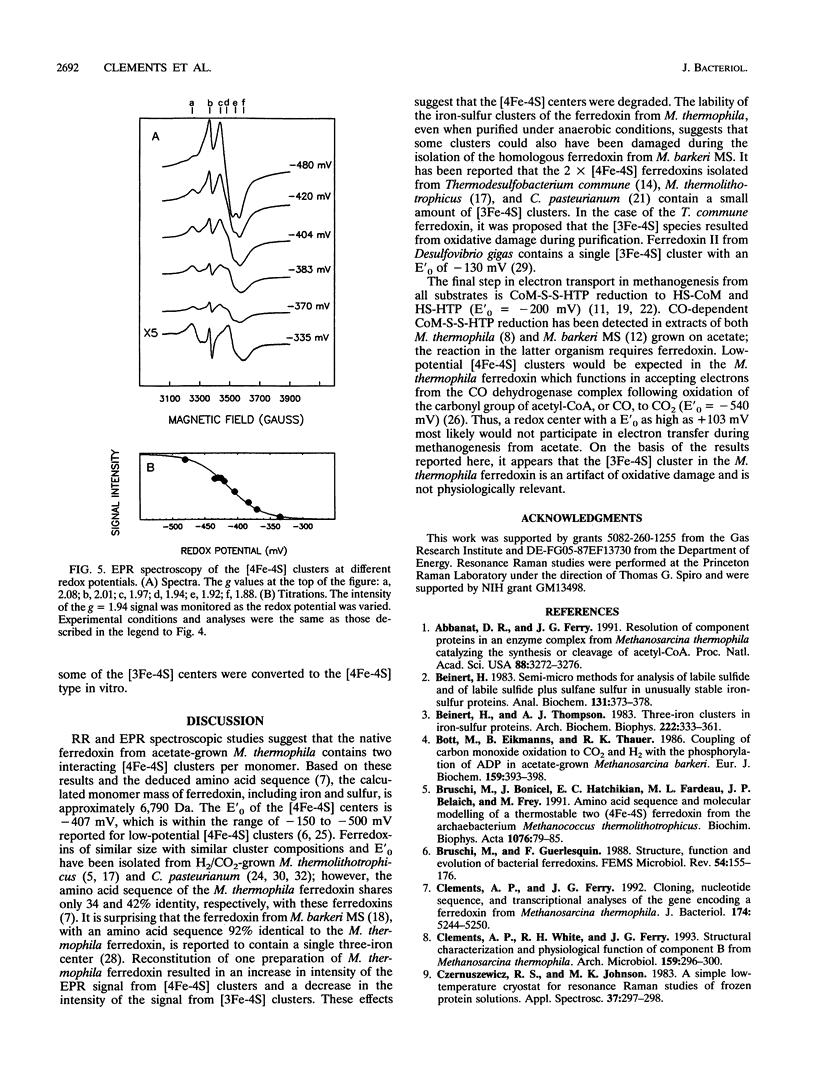
Abstract
Ferredoxin from Methanosarcina thermophila is an electron acceptor for the CO dehydrogenase complex which decarbonylates acetyl-coenzyme A and oxidizes the carbonyl group to carbon dioxide in the pathway for conversion of the methyl group of acetate to methane (K. C. Terlesky and J. G. Ferry, J. Biol. Chem. 263:4080-4082, 1988). Resonance Raman spectroscopy and electron paramagnetic resonance spectroelectrochemistry indicated that the ferredoxin contained two [4Fe-4S] clusters per monomer of 6,790 Da, each with a midpoint potential of -407 mV. A [3Fe-4S] species, with a midpoint potential of +103 mV, was also detected in the protein at high redox potentials. Quantitation of the [3Fe-4S] and [4Fe-4S] centers revealed 0.4 and 2.1 spins per monomer, respectively. The iron-sulfur clusters were unstable in the presence of air, and the rate of cluster loss increased with increasing temperature. A ferredoxin preparation, with a low spin quantitation of [4Fe-4S] centers, was treated with Fe2+ and S2-, which resulted in an increase in [4Fe-4S] and a decrease in [3Fe-4S] clusters. The results of these studies suggest the [3Fe-4S] species may be an artifact formed from degradation of [4Fe-4S] clusters.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abbanat D. R., Ferry J. G. Resolution of component proteins in an enzyme complex from Methanosarcina thermophila catalyzing the synthesis or cleavage of acetyl-CoA. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3272–3276. doi: 10.1073/pnas.88.8.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983 Jun;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- Beinert H., Thomson A. J. Three-iron clusters in iron-sulfur proteins. Arch Biochem Biophys. 1983 Apr 15;222(2):333–361. doi: 10.1016/0003-9861(83)90531-3. [DOI] [PubMed] [Google Scholar]
- Bott M., Eikmanns B., Thauer R. K. Coupling of carbon monoxide oxidation to CO2 and H2 with the phosphorylation of ADP in acetate-grown Methanosarcina barkeri. Eur J Biochem. 1986 Sep 1;159(2):393–398. doi: 10.1111/j.1432-1033.1986.tb09881.x. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Bonicel J., Hatchikian E. C., Fardeau M. L., Belaich J. P., Frey M. Amino acid sequence and molecular modelling of a thermostable two (4Fe-4S) ferredoxin from the archaebacterium Methanococcus thermolithotrophicus. Biochim Biophys Acta. 1991 Jan 8;1076(1):79–85. doi: 10.1016/0167-4838(91)90222-l. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Guerlesquin F. Structure, function and evolution of bacterial ferredoxins. FEMS Microbiol Rev. 1988 Apr-Jun;4(2):155–175. doi: 10.1111/j.1574-6968.1988.tb02741.x. [DOI] [PubMed] [Google Scholar]
- Clements A. P., Ferry J. G. Cloning, nucleotide sequence, and transcriptional analyses of the gene encoding a ferredoxin from Methanosarcina thermophila. J Bacteriol. 1992 Aug;174(16):5244–5250. doi: 10.1128/jb.174.16.5244-5250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R., Thauer R. K. Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS). FEBS Lett. 1990 Sep 3;269(2):368–372. doi: 10.1016/0014-5793(90)81195-t. [DOI] [PubMed] [Google Scholar]
- Fish W. W. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- Harder S. R., Lu W. P., Feinberg B. A., Ragsdale S. W. Spectroelectrochemical studies of the corrinoid/iron-sulfur protein involved in acetyl coenzyme A synthesis by Clostridium thermoaceticum. Biochemistry. 1989 Nov 14;28(23):9080–9087. doi: 10.1021/bi00449a019. [DOI] [PubMed] [Google Scholar]
- Hatchikian E. C., Bruschi M., Forget N., Scandellari M. Electron transport components from methanogenic bacteria: the ferredoxin from Methanosarcina barkeri (strain Fusaro). Biochem Biophys Res Commun. 1982 Dec 31;109(4):1316–1323. doi: 10.1016/0006-291x(82)91921-0. [DOI] [PubMed] [Google Scholar]
- Hatchikian E. C., Fardeau M. L., Bruschi M., Belaich J. P., Chapman A., Cammack R. Isolation, characterization, and biological activity of the Methanococcus thermolithotrophicus ferredoxin. J Bacteriol. 1989 May;171(5):2384–2390. doi: 10.1128/jb.171.5.2384-2390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausinger R. P., Moura I., Moura J. J., Xavier A. V., Santos M. H., LeGall J., Howard J. B. Amino acid sequence of a 3Fe:3S ferredoxin from the "archaebacterium" Methanosarcina barkeri (DSM 800). J Biol Chem. 1982 Dec 10;257(23):14192–14197. [PubMed] [Google Scholar]
- Johnson M. K., Spiro T. G., Mortenson L. E. Resonance Raman and electron paramagnetic resonance studies on oxidized and ferricyanide-treated Clostridium pasteurianum ferredoxin. Vibrational assignments from 34S shifts and evidence for conversion of 4 to 3 iron-sulfur clusters via oxidative damage. Vibrational assignments from 34S shifts and evidence for conversion of 4 to 3 iron-sulfur clusters via oxidative damage. J Biol Chem. 1982 Mar 10;257(5):2447–2452. [PubMed] [Google Scholar]
- Kell D. B., Morris J. G. Oxidation-reduction properties of coenzyme M (2-mercaptoethane sulphonate) at the mercury electrode. FEBS Lett. 1979 Dec 15;108(2):481–484. doi: 10.1016/0014-5793(79)80593-1. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mathews R., Charlton S., Sands R. H., Palmer G. On the nature of the spin coupling between the iron-sulfur clusters in the eight-iron ferredoxins. J Biol Chem. 1974 Jul 10;249(13):4326–4328. [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Moura I., Moura J. J., Huynh B. H., Santos H., LeGall J., Xavier A. V. Ferredoxin from Methanosarcina barkeri: evidence for the presence of a three-iron center. Eur J Biochem. 1982 Aug;126(1):95–98. doi: 10.1111/j.1432-1033.1982.tb06751.x. [DOI] [PubMed] [Google Scholar]
- Moura J. J., LeGall J., Xavier A. V. Interconversion from 3Fe into 4Fe clusters in the presence of Desulfovibrio gigas cell extracts. Eur J Biochem. 1984 Jun 1;141(2):319–322. doi: 10.1111/j.1432-1033.1984.tb08194.x. [DOI] [PubMed] [Google Scholar]
- Prince R. C., Adams M. W. Oxidation-reduction properties of the two Fe4S4 clusters in Clostridium pasteurianum ferredoxin. J Biol Chem. 1987 Apr 15;262(11):5125–5128. [PubMed] [Google Scholar]
- Tanaka M., Nakashima T., Benson A., Mower H., Tasunobu K. T. The amino acid sequence of Clostridium pasteurianum ferredoxin. Biochemistry. 1966 May;5(5):1666–1681. doi: 10.1021/bi00869a032. [DOI] [PubMed] [Google Scholar]
- Terlesky K. C., Ferry J. G. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988 Mar 25;263(9):4075–4079. [PubMed] [Google Scholar]
- Terlesky K. C., Ferry J. G. Purification and characterization of a ferredoxin from acetate-grown Methanosarcina thermophila. J Biol Chem. 1988 Mar 25;263(9):4080–4082. [PubMed] [Google Scholar]


