Abstract
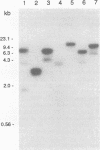
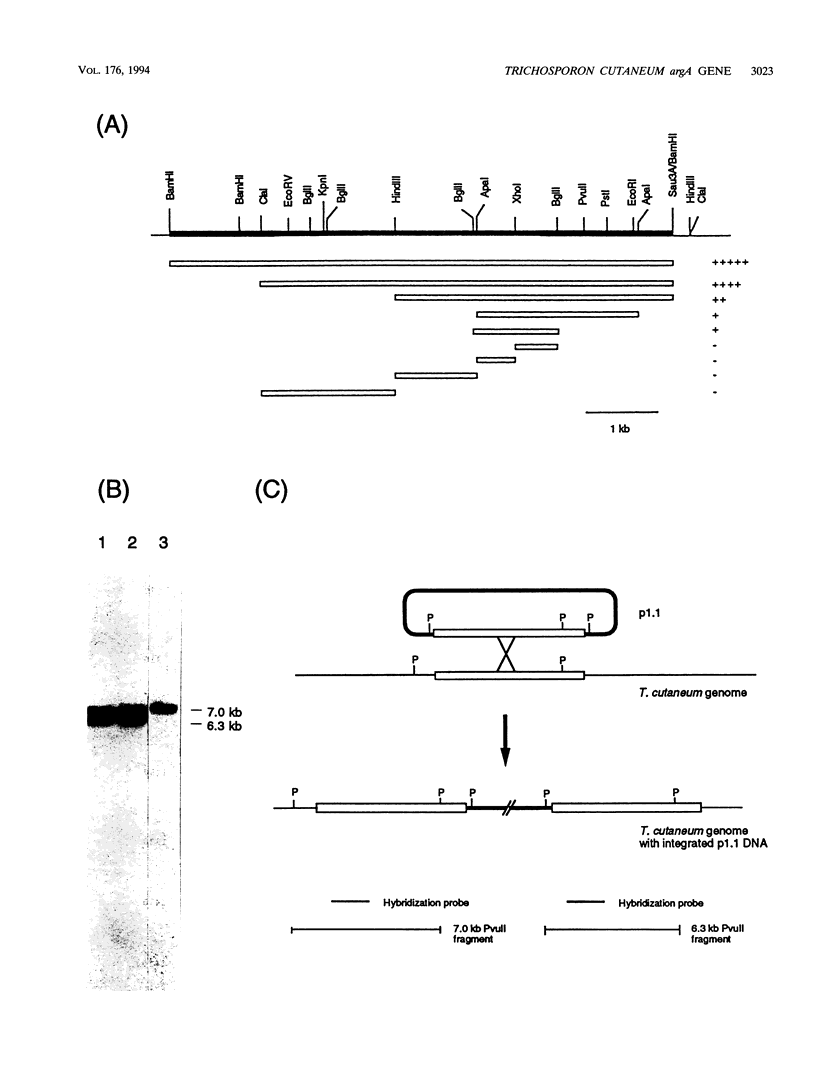
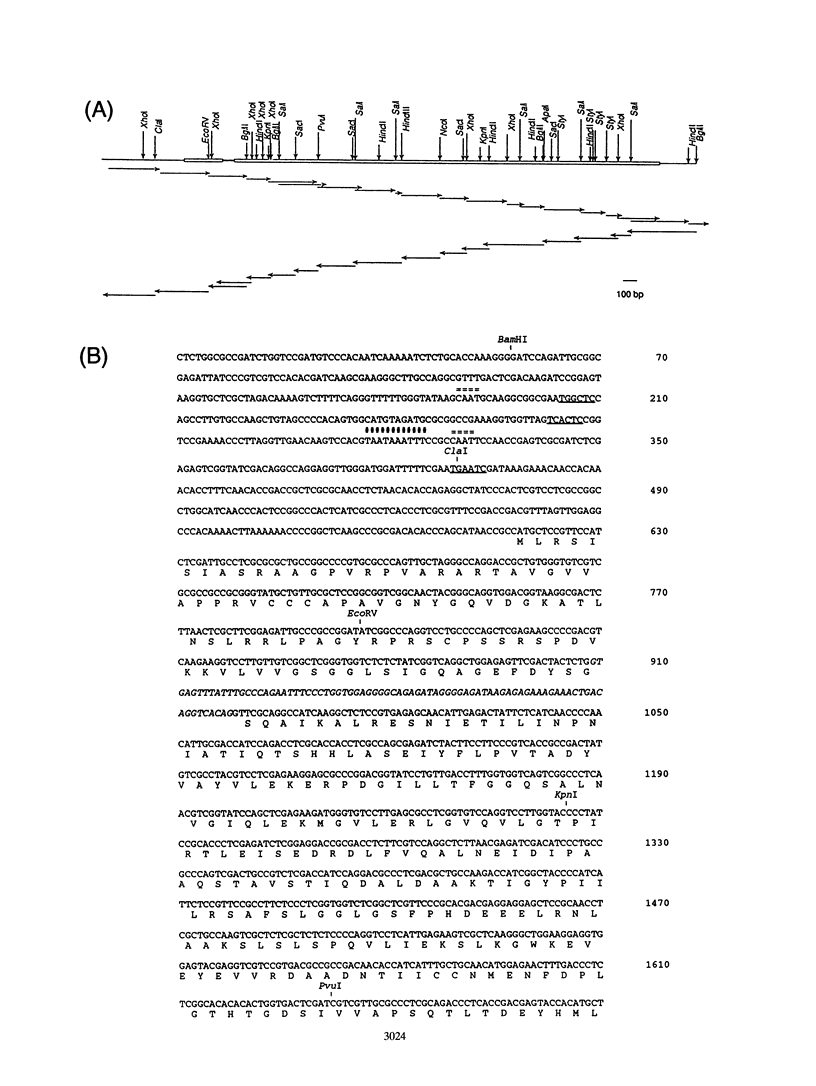
Genomic clones capable of complementing a previously isolated arginine auxotrophic mutant strain of the filamentous yeast Trichosporon cutaneum DSM 70698 have been identified by DNA-mediated transformation, and a complementing 4,082-bp subfragment was sequenced. This analysis revealed an intact gene (arg4) showing a high degree of homology with the Saccharomyces cerevisiae CPA2 gene encoding the large subunit of carbamoyl-phosphate synthetase (CPS-A). The inferred amino acid sequence of the T. cutaneum argA-encoded protein contains 1,168 residues showing 62% identity with the sequence of the S. cerevisiae CPA2 protein, and the comparison of the two sequences uncovered a putative intron sequence of 81 nucleotides close to the 5' end of the coding region of the T. cutaneum argA gene. The presence of this intron was confirmed by nuclease protection studies and by direct DNA sequence analysis of a cDNA fragment which had been obtained by PCR amplification. The T. cutaneum intron shares the general characteristics of introns found in yeasts and filamentous fungi. A major transcript of around 4 kb was found in Northern (RNA) blots. The T. cutaneum argA coding region was expressed in Escherichia coli under the control of the regulatable tac promoter. A roughly 130-kDa protein which was found to cross-react with an anti-rat CPS antibody in Western blots (immunoblots) was observed. Two putative ATP-binding domains were identified, one in the amino-terminal half of the argA-encoded protein and the other in the carboxy-terminal half. These domains are highly conserved among the known CPS-A sequences from S. cerevisiae, E. coli, and the rat. From these results we conclude that the T. cutaneum argA gene encodes the large subunit of CPS. This is the first gene to be identified and analyzed in the T. cutaneum DSM 70698 strain.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Ochs B., Abel K. J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988 Sep 30;69(2):301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Bernhardt S. A., Davis R. H. Carbamoyl phosphate compartmentation in Neurospora: histochemical localization of aspartate and ornithine transcarbamoylases. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1868–1872. doi: 10.1073/pnas.69.7.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman B. J., Davis R. H. Cellular distribution of ornithine in Neurospora: anabolic and catabolic steady states. J Bacteriol. 1977 Apr;130(1):274–284. doi: 10.1128/jb.130.1.274-284.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H. Metabolite distribution in cells. Science. 1972 Nov 24;178(4063):835–840. doi: 10.1126/science.178.4063.835. [DOI] [PubMed] [Google Scholar]
- Davis R. H., Ristow J. L., Hanson B. A. Carbamyl phosphate synthetase A of Neurospora crassa. J Bacteriol. 1980 Jan;141(1):144–155. doi: 10.1128/jb.141.1.144-155.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mot R., Verachtert H. Secretion of alpha-amylase and multiple forms of glucoamylase by the yeast Trichosporon pullulans. Can J Microbiol. 1986 Jan;32(1):47–51. doi: 10.1139/m86-009. [DOI] [PubMed] [Google Scholar]
- Devaney M. A., Powers-Lee S. G. Immunological cross-reactivity between carbamyl phosphate synthetases I, II, and III. J Biol Chem. 1984 Jan 25;259(2):703–706. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman J. C., Kwon-Chung K. J. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990 Sep;10(9):4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gigot D., Crabeel M., Feller A., Charlier D., Lissens W., Glansdorff N., Piérard A. Patterns of polarity in the Escherichia coli car AB gene cluster. J Bacteriol. 1980 Aug;143(2):914–920. doi: 10.1128/jb.143.2.914-920.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glumoff V., Käppeli O., Fiechter A., Reiser J. Genetic transformation of the filamentous yeast, Trichosporon cutaneum, using dominant selection markers. Gene. 1989 Dec 14;84(2):311–318. doi: 10.1016/0378-1119(89)90505-2. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guého E., Smith M. T., de Hoog G. S., Billon-Grand G., Christen R., Batenburg-van der Vegte W. H. Contributions to a revision of the genus Trichosporon. Antonie Van Leeuwenhoek. 1992 May;61(4):289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- Guého E., Tredick J., Phaff H. J. DNA base composition and DNA relatedness among species of Trichosporon Behrend. Antonie Van Leeuwenhoek. 1984;50(1):17–32. doi: 10.1007/BF00404904. [DOI] [PubMed] [Google Scholar]
- Guého E., de Hoog G. S., Smith M. T. Neotypification of the genus Trichosporon. Antonie Van Leeuwenhoek. 1992 May;61(4):285–288. doi: 10.1007/BF00713937. [DOI] [PubMed] [Google Scholar]
- Hawkes R., Niday E., Gordon J. A dot-immunobinding assay for monoclonal and other antibodies. Anal Biochem. 1982 Jan 1;119(1):142–147. doi: 10.1016/0003-2697(82)90677-7. [DOI] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Hoy J., Hsu K. C., Rolston K., Hopfer R. L., Luna M., Bodey G. P. Trichosporon beigelii infection: a review. Rev Infect Dis. 1986 Nov-Dec;8(6):959–967. [PubMed] [Google Scholar]
- Hug H., Blanch H. W., Fiechter A. The functional role of lipids in hydrocarbon assimilation. Biotechnol Bioeng. 1974 Jul;16(7):965–985. doi: 10.1002/bit.260160709. [DOI] [PubMed] [Google Scholar]
- Karlin J. N., Bowman B. J., Davis R. H. Compartmental behavior of ornithine in Neurospora crassa. J Biol Chem. 1976 Jul 10;251(13):3948–3955. [PubMed] [Google Scholar]
- Kemker B. J., Lehmann P. F., Lee J. W., Walsh T. J. Distinction of deep versus superficial clinical and nonclinical isolates of Trichosporon beigelii by isoenzymes and restriction fragment length polymorphisms of rDNA generated by polymerase chain reaction. J Clin Microbiol. 1991 Aug;29(8):1677–1683. doi: 10.1128/jcm.29.8.1677-1683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A. Improved synthesis of full-length RNA probe at reduced incubation temperatures. Nucleic Acids Res. 1990 Nov 11;18(21):6463–6463. doi: 10.1093/nar/18.21.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kälin M., Neujahr H. Y., Weissmahr R. N., Sejlitz T., Jöhl R., Fiechter A., Reiser J. Phenol hydroxylase from Trichosporon cutaneum: gene cloning, sequence analysis, and functional expression in Escherichia coli. J Bacteriol. 1992 Nov;174(22):7112–7120. doi: 10.1128/jb.174.22.7112-7120.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroute F., Piérard A., Grenson M., Wiame J. M. The biosynthesis of carbamoyl phosphate in Saccharomyces cerevisiae. J Gen Microbiol. 1965 Jul;40(1):127–142. doi: 10.1099/00221287-40-1-127. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Lee J. W., Melcher G. A., Rinaldi M. G., Pizzo P. A., Walsh T. J. Patterns of morphologic variation among isolates of Trichosporon beigelii. J Clin Microbiol. 1990 Dec;28(12):2823–2827. doi: 10.1128/jcm.28.12.2823-2827.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusty C. J., Widgren E. E., Broglie K. E., Nyunoya H. Yeast carbamyl phosphate synthetase. Structure of the yeast gene and homology to Escherichia coli carbamyl phosphate synthetase. J Biol Chem. 1983 Dec 10;258(23):14466–14477. [PubMed] [Google Scholar]
- Makoff A. J., Radford A. Genetics and biochemistry of carbamoyl phosphate biosynthesis and its utilization in the pyrimidine biosynthetic pathway. Microbiol Rev. 1978 Jun;42(2):307–328. doi: 10.1128/mr.42.2.307-328.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay M., Gigot D., Beckmann J., Glansdorff N., Piérard A. Physiology and genetics of carbamoylphosphate synthesis in Escherichia coli K12. Mol Gen Genet. 1974;133(4):299–316. doi: 10.1007/BF00332706. [DOI] [PubMed] [Google Scholar]
- Mertins P., Gallwitz D. Nuclear pre-mRNA splicing in the fission yeast Schizosaccharomyces pombe strictly requires an intron-contained, conserved sequence element. EMBO J. 1987 Jun;6(6):1757–1763. doi: 10.1002/j.1460-2075.1987.tb02428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr H. Y. Yeasts in biodegradation and biodeterioration processes. Bioprocess Technol. 1990;5:321–348. [PubMed] [Google Scholar]
- Nyunoya H., Broglie K. E., Lusty C. J. The gene coding for carbamoyl-phosphate synthetase I was formed by fusion of an ancestral glutaminase gene and a synthetase gene. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2244–2246. doi: 10.1073/pnas.82.8.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyunoya H., Broglie K. E., Widgren E. E., Lusty C. J. Characterization and derivation of the gene coding for mitochondrial carbamyl phosphate synthetase I of rat. J Biol Chem. 1985 Aug 5;260(16):9346–9356. [PubMed] [Google Scholar]
- Nyunoya H., Lusty C. J. Sequence of the small subunit of yeast carbamyl phosphate synthetase and identification of its catalytic domain. J Biol Chem. 1984 Aug 10;259(15):9790–9798. [PubMed] [Google Scholar]
- Nyunoya H., Lusty C. J. The carB gene of Escherichia coli: a duplicated gene coding for the large subunit of carbamoyl-phosphate synthetase. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4629–4633. doi: 10.1073/pnas.80.15.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U. A., Glumoff V., Kälin M., Fiechter A., Reiser J. Genetic transformation of auxotrophic mutants of the filamentous yeast Trichosporon cutaneum using homologous and heterologous marker genes. Yeast. 1991 Jul;7(5):513–524. doi: 10.1002/yea.320070511. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Sachs M. S., Yanofsky C. The Neurospora crassa arg-2 locus. Structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J Biol Chem. 1990 Jul 5;265(19):10981–10987. [PubMed] [Google Scholar]
- Piérard A., Schröter B. Structure-function relationships in the arginine pathway carbamoylphosphate synthase of Saccharomyces cerevisiae. J Bacteriol. 1978 Apr;134(1):167–176. doi: 10.1128/jb.134.1.167-176.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser J., Stark G. R. Immunologic detection of specific proteins in cell extracts by fractionation in gels and transfer to paper. Methods Enzymol. 1983;96:205–215. doi: 10.1016/s0076-6879(83)96018-4. [DOI] [PubMed] [Google Scholar]
- Reiser J., Walther I. S., Fraefel C., Fiechter A. Methods to investigate the expression of lignin peroxidase genes by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1993 Sep;59(9):2897–2903. doi: 10.1128/aem.59.9.2897-2903.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino S. D., Nyunoya H., Lusty C. J. Catalytic domains of carbamyl phosphate synthetase. Glutamine-hydrolyzing site of Escherichia coli carbamyl phosphate synthetase. J Biol Chem. 1986 Aug 25;261(24):11320–11327. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer J. P., Kelly R. E., Rinker A. G., Jr, Scully J. L., Evans D. R. Mammalian carbamyl phosphate synthetase (CPS). DNA sequence and evolution of the CPS domain of the Syrian hamster multifunctional protein CAD. J Biol Chem. 1990 Jun 25;265(18):10395–10402. [PubMed] [Google Scholar]
- Skrzypek M., Borsuk P., Maleszka R. Cloning and sequencing of the ornithine carbamoyltransferase gene from Pachysolen tannophilus. Yeast. 1990 Mar-Apr;6(2):141–148. doi: 10.1002/yea.320060208. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sze I. S., Dagley S. Properties of salicylate hydroxylase and hydroxyquinol 1,2-dioxygenase purified from Trichosporon cutaneum. J Bacteriol. 1984 Jul;159(1):353–359. doi: 10.1128/jb.159.1.353-359.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeri T. T., Kumar V., Lehtovaara P., Knowles J. Construction of cDNA libraries by blunt-end ligation: high-frequency cloning of long cDNAs from filamentous fungi. Anal Biochem. 1987 Jul;164(1):60–67. doi: 10.1016/0003-2697(87)90367-8. [DOI] [PubMed] [Google Scholar]
- Upshall A., Gilbert T., Saari G., O'Hara P. J., Weglenski P., Berse B., Miller K., Timberlake W. E. Molecular analysis of the argB gene of Aspergillus nidulans. Mol Gen Genet. 1986 Aug;204(2):349–354. doi: 10.1007/BF00425521. [DOI] [PubMed] [Google Scholar]
- Urrestarazu L. A., Vissers S., Wiame J. M. Change in location of ornithine carbamoyltransferase and carbamoylphosphate synthetase among yeasts in relation to the arginase/ornithine carbamoyltransferase regulatory complex and the energy status of the cells. Eur J Biochem. 1977 Oct 3;79(2):473–481. doi: 10.1111/j.1432-1033.1977.tb11830.x. [DOI] [PubMed] [Google Scholar]
- Van Der Walt J. P., Hopsu-Havu V. K. A colour reaction for the differentiation of ascomycetous and hemibasidiomycetous yeasts. Antonie Van Leeuwenhoek. 1976;42(1-2):157–163. doi: 10.1007/BF00399460. [DOI] [PubMed] [Google Scholar]
- Walsh T. J., Melcher G. P., Lee J. W., Pizzo P. A. Infections due to Trichosporon species: new concepts in mycology, pathogenesis, diagnosis and treatment. Curr Top Med Mycol. 1993;5:79–113. [PubMed] [Google Scholar]
- Ward M., Wilkinson B., Turner G. Transformation of Aspergillus nidulans with a cloned, oligomycin-resistant ATP synthase subunit 9 gene. Mol Gen Genet. 1986 Feb;202(2):265–270. doi: 10.1007/BF00331648. [DOI] [PubMed] [Google Scholar]
- Weiss R. L., Davis R. H. Intracellular localization of enzymes of arginine metabolism in Neurospora. J Biol Chem. 1973 Aug 10;248(15):5403–5408. [PubMed] [Google Scholar]
- West M., Emerson G. W., Sullivan P. A. Purification and properties of two lactose hydrolases from Trichosporon cutaneum. J Gen Microbiol. 1990 Aug;136(8):1483–1490. doi: 10.1099/00221287-136-8-1483. [DOI] [PubMed] [Google Scholar]
- Williams L. G., Bernhardt S. A., Davis R. H. Evidence for two discrete carbamyl phosphate pools in Neurospora. J Biol Chem. 1971 Feb 25;246(4):973–978. [PubMed] [Google Scholar]
- van Gorcom R. F., Punt P. J., Pouwels P. H., van den Hondel C. A. A system for the analysis of expression signals in Aspergillus. Gene. 1986;48(2-3):211–217. doi: 10.1016/0378-1119(86)90079-x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986 Jun;5(6):1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G., Steppuhn J., Herrmann R. G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989 Apr 1;180(3):535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]