Abstract
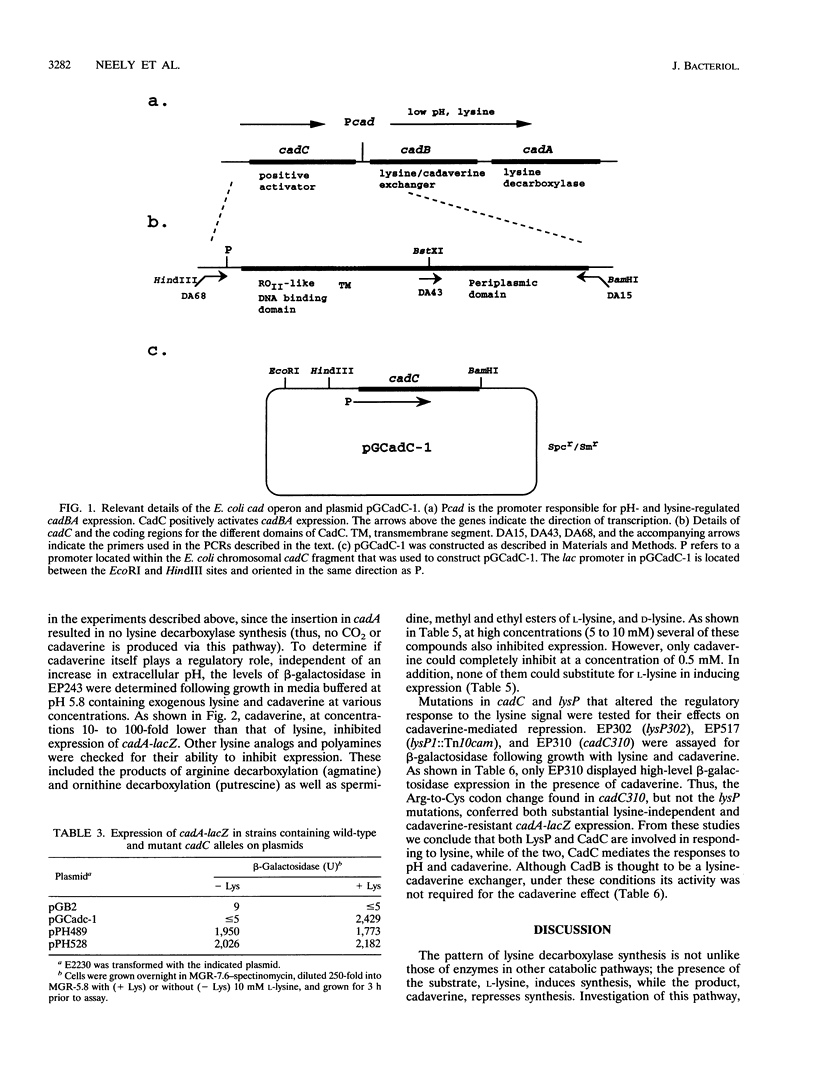
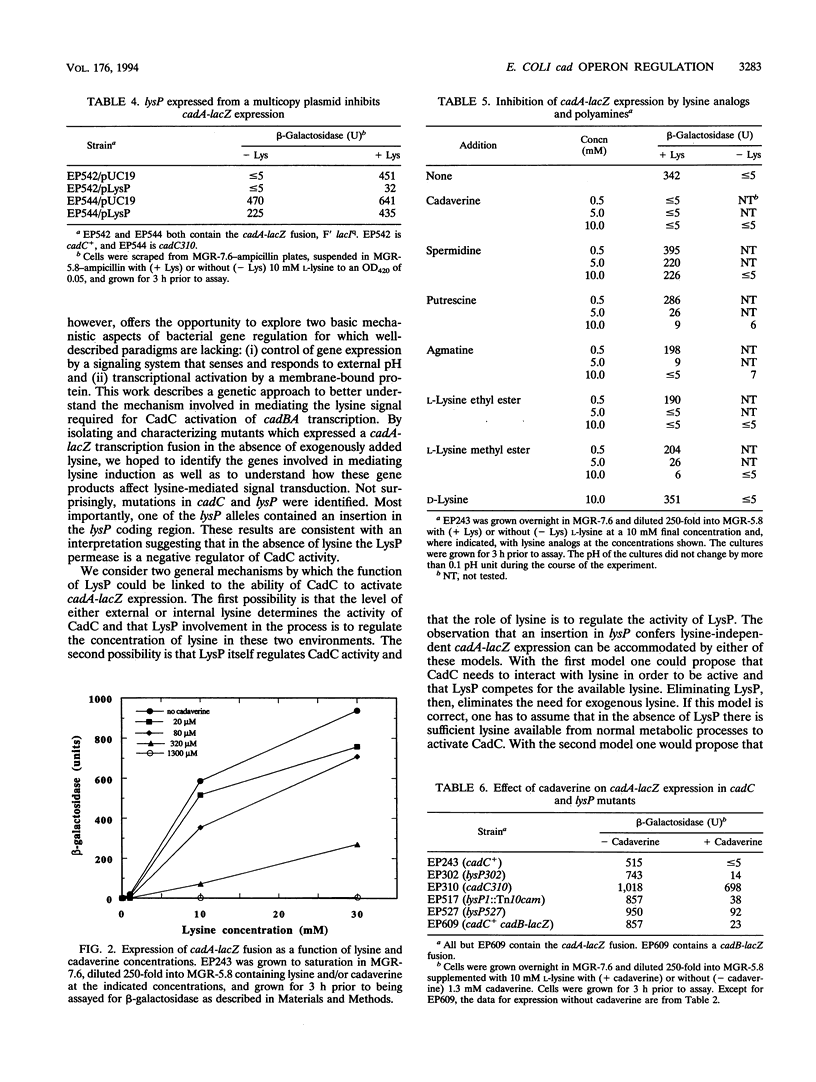
Expression of the Escherichia coli cadBA operon, encoding functions required for the conversion of lysine to cadaverine and for cadaverine excretion, requires at least two extracellular signals: low pH and a high concentration of lysine. To better understand the nature of the lysine-dependent signal, mutants were isolated which expressed a cadA-lacZ transcription fusion in the absence of lysine while retaining pH regulation. The responsible mutation in one of these isolates (EP310) was in cadC, a gene encoding a function necessary for transcriptional activation of cadBA. This mutation (cadC310) is in a part of the gene encoding the periplasmic domain of CadC and results in an Arg-to-Cys change at position 265, indicating that this part of the protein is involved in responding to the presence of lysine. Three other mutants had mutations mapping in or near lysP (cadR), a gene encoding a lysine transport protein that has previously been shown to regulate cadA expression. One of these mutations is an insertion in the lysP coding region. Thus, in the absence of exogenous lysine, LysP is a negative regulator of cadBA expression. Negative regulation by LysP was further demonstrated by showing that lysP expression from a high-copy-number plasmid rendered cadA-lacZ uninducible. Expression of cadA-lacZ in a strain carrying the cadC310 allele, however, was not affected by the plasmid-expressed lysP. Cadaverine was shown to inhibit expression of the cadA-lacZ fusion in cadC+ cells but not in a cadC310 background.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auger E. A., Redding K. E., Plumb T., Childs L. C., Meng S. Y., Bennett G. N. Construction of lac fusions to the inducible arginine- and lysine decarboxylase genes of Escherichia coli K12. Mol Microbiol. 1989 May;3(5):609–620. doi: 10.1111/j.1365-2958.1989.tb00208.x. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth I. R. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985 Dec;49(4):359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- DiRita V. J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992 Feb;6(4):451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Ehrmann M., Boos W. Identification of endogenous inducers of the mal regulon in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3539–3545. doi: 10.1128/jb.169.8.3539-3545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Hall H. K. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Hall H. K. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J Bacteriol. 1991 Aug;173(16):5129–5135. doi: 10.1128/jb.173.16.5129-5135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J Bacteriol. 1991 Nov;173(21):6896–6902. doi: 10.1128/jb.173.21.6896-6902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W. The acid tolerance response of Salmonella typhimurium involves transient synthesis of key acid shock proteins. J Bacteriol. 1993 Apr;175(7):1981–1987. doi: 10.1128/jb.175.7.1981-1987.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti A. A., Krulwich T. A. Bioenergetic problems of alkalophilic bacteria. Biochem Soc Trans. 1984 Jun;12(3):411–412. doi: 10.1042/bst0120411. [DOI] [PubMed] [Google Scholar]
- Joshi A. K., Baichwal V., Ames G. F. Rapid polymerase chain reaction amplification using intact bacterial cells. Biotechniques. 1991 Jan;10(1):42, 44-5. [PubMed] [Google Scholar]
- Kleckner N., Bender J., Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Kroll R. G., Booth I. R. The relationship between intracellular pH, the pH gradient and potassium transport in Escherichia coli. Biochem J. 1983 Dec 15;216(3):709–716. doi: 10.1042/bj2160709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krulwich T. A., Guffanti A. A. Alkalophilic bacteria. Annu Rev Microbiol. 1989;43:435–463. doi: 10.1146/annurev.mi.43.100189.002251. [DOI] [PubMed] [Google Scholar]
- Kühnau S., Reyes M., Sievertsen A., Shuman H. A., Boos W. The activities of the Escherichia coli MalK protein in maltose transport, regulation, and inducer exclusion can be separated by mutations. J Bacteriol. 1991 Apr;173(7):2180–2186. doi: 10.1128/jb.173.7.2180-2186.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S. Y., Bennett G. N. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol. 1992 Apr;174(8):2659–2669. doi: 10.1128/jb.174.8.2659-2669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng S. Y., Bennett G. N. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J Bacteriol. 1992 Apr;174(8):2670–2678. doi: 10.1128/jb.174.8.2670-2678.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E. R. Influence of pH on bacterial gene expression. Mol Microbiol. 1993 Apr;8(1):5–14. doi: 10.1111/j.1365-2958.1993.tb01198.x. [DOI] [PubMed] [Google Scholar]
- Padan E., Schuldiner S. Intracellular pH and membrane potential as regulators in the prokaryotic cell. J Membr Biol. 1987;95(3):189–198. doi: 10.1007/BF01869481. [DOI] [PubMed] [Google Scholar]
- Padan E., Schuldiner S. Intracellular pH regulation in bacterial cells. Methods Enzymol. 1986;125:337–352. doi: 10.1016/s0076-6879(86)25029-6. [DOI] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Popkin P. S., Maas W. K. Escherichia coli regulatory mutation affecting lysine transport and lysine decarboxylase. J Bacteriol. 1980 Feb;141(2):485–492. doi: 10.1128/jb.141.2.485-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja N., Goodson M., Chui W. C., Smith D. G., Rowbury R. J. Habituation to acid in Escherichia coli: conditions for habituation and its effects on plasmid transfer. J Appl Bacteriol. 1991 Jan;70(1):59–65. doi: 10.1111/j.1365-2672.1991.tb03787.x. [DOI] [PubMed] [Google Scholar]
- Reyes M., Shuman H. A. Overproduction of MalK protein prevents expression of the Escherichia coli mal regulon. J Bacteriol. 1988 Oct;170(10):4598–4602. doi: 10.1128/jb.170.10.4598-4602.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowbury R. J., Goodson M. PhoE porin of Escherichia coli and phosphate reversal of acid damage and killing and of acid induction of the CadA gene product. J Appl Bacteriol. 1993 Jun;74(6):652–661. doi: 10.1111/j.1365-2672.1993.tb05199.x. [DOI] [PubMed] [Google Scholar]
- Rowbury R. J., Goodson M., Wallace A. D. The PhoE porin and transmission of the chemical stimulus for induction of acid resistance (acid habituation) in Escherichia coli. J Appl Bacteriol. 1992 Mar;72(3):233–243. doi: 10.1111/j.1365-2672.1992.tb01829.x. [DOI] [PubMed] [Google Scholar]
- Sabo D. L., Boeker E. A., Byers B., Waron H., Fischer E. H. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. Biochemistry. 1974 Feb 12;13(4):662–670. doi: 10.1021/bi00701a005. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Shi X., Waasdorp B. C., Bennett G. N. Modulation of acid-induced amino acid decarboxylase gene expression by hns in Escherichia coli. J Bacteriol. 1993 Feb;175(4):1182–1186. doi: 10.1128/jb.175.4.1182-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M., Baker T. A., Schnitzler G., Deischel S. M., Goel M., Dove W., Jaacks K. J., Grossman A. D., Erickson J. W., Gross C. A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989 Mar;53(1):1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slonczewski J. L., Gonzalez T. N., Bartholomew F. M., Holt N. J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987 Jul;169(7):3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffes C., Ellis J., Wu J., Rosen B. P. The lysP gene encodes the lysine-specific permease. J Bacteriol. 1992 May;174(10):3242–3249. doi: 10.1128/jb.174.10.3242-3249.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor H., Hafner E. W., Tabor C. W. Construction of an Escherichia coli strain unable to synthesize putrescine, spermidine, or cadaverine: characterization of two genes controlling lysine decarboxylase. J Bacteriol. 1980 Dec;144(3):952–956. doi: 10.1128/jb.144.3.952-956.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L., Kodaira R., Neidhardt F. C. Physiological regulation of a decontrolled lac operon. J Bacteriol. 1977 Apr;130(1):212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N., Dunyak D. S., Rosey E. L., Slonczewski J. L., Olson E. R. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol. 1992 Jan;174(2):530–540. doi: 10.1128/jb.174.2.530-540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev. 1992 Mar;56(1):12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]



