Abstract
Crude extracts from Escherichia coli were screened for any broad-specificity endoribonuclease after the cell proteins were fractionated by size. In a mutant lacking the gene for RNase I (molecular mass, 27,156 Da), the only such activities were also in the size range of 23 to 28 kDa. Fractionation by chromatography on a strong cation-exchange resin revealed only two activities. One of them eluted at a salt concentration expected for RNase M and had the specificity of RNase M. It preferred pyrimidine-adenosine bonds, could not degrade purine homopolymers, and had a molecular mass of approximately 27 kDa (V. J. Cannistraro and D. Kennell, Eur. J. Biochem. 181:363-370, 1989). A second fraction, eluting at a higher salt concentration, was active against any phosphodiester bond but was about 100 times less active than are RNase I and RNase I* (a form of RNase I) in the wild-type cell. On the basis of sizing-gel chromatography, this enzyme had a molecular mass of approximately 24 kDa. We call it RNase R (for residual). RNase R is not an abnormal product of the mutant rna gene; a cell carrying many copies of that gene on a plasmid did not synthesize more RNase R. Our search for broad-specificity endoribonucleases was prompted by the expectation that the primary activities for mRNA degradation are expressed by a relatively small number of broad-specificity RNases. If correct, the results suggest that the endoribonucleases for this major metabolic activity reside in the 24- to 28-kDa size range. Endoribonucleases with much greater specificity must have as primary functions the processing of specific RNA molecules at a very limited number of sites as steps in their biosynthesis. In exceptional cases, these endoribonucleases inactivate a specific message that has such a site, and they can also effect total mRNA metabolism indirectly by a global disturbance of the cell physiology. It is suggested that a distinction be made between these processing and degradative activities.
Full text
PDF
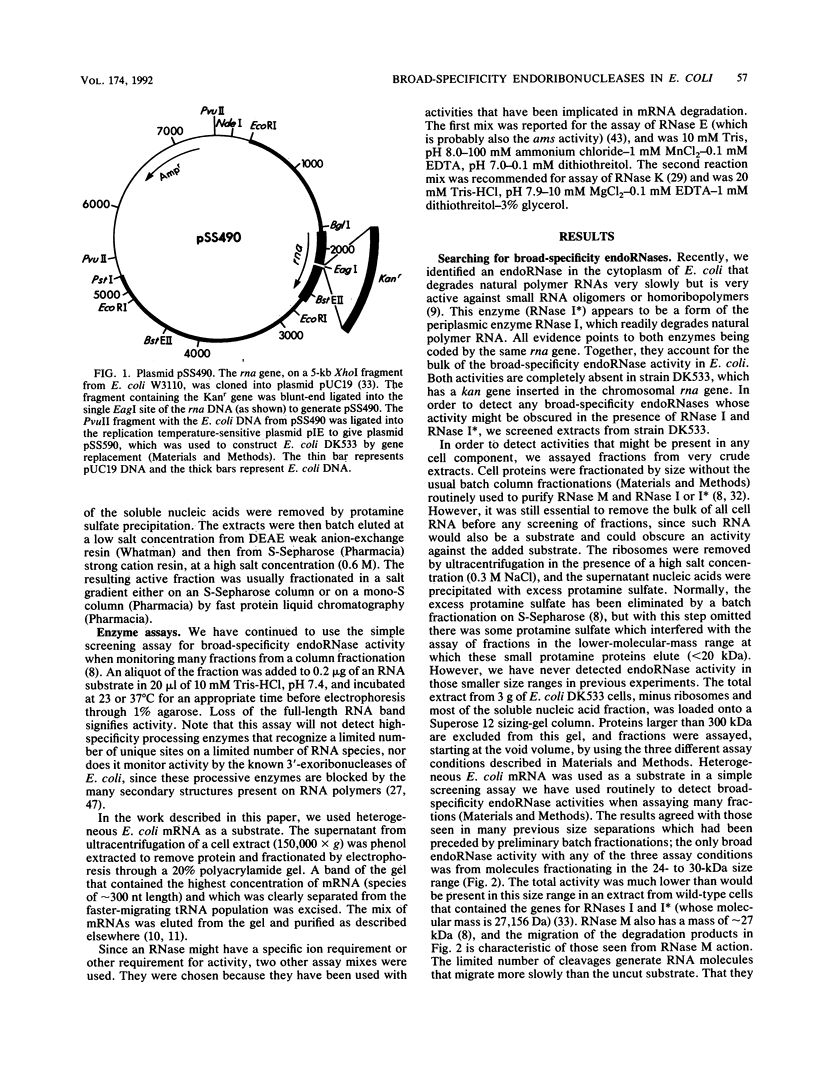
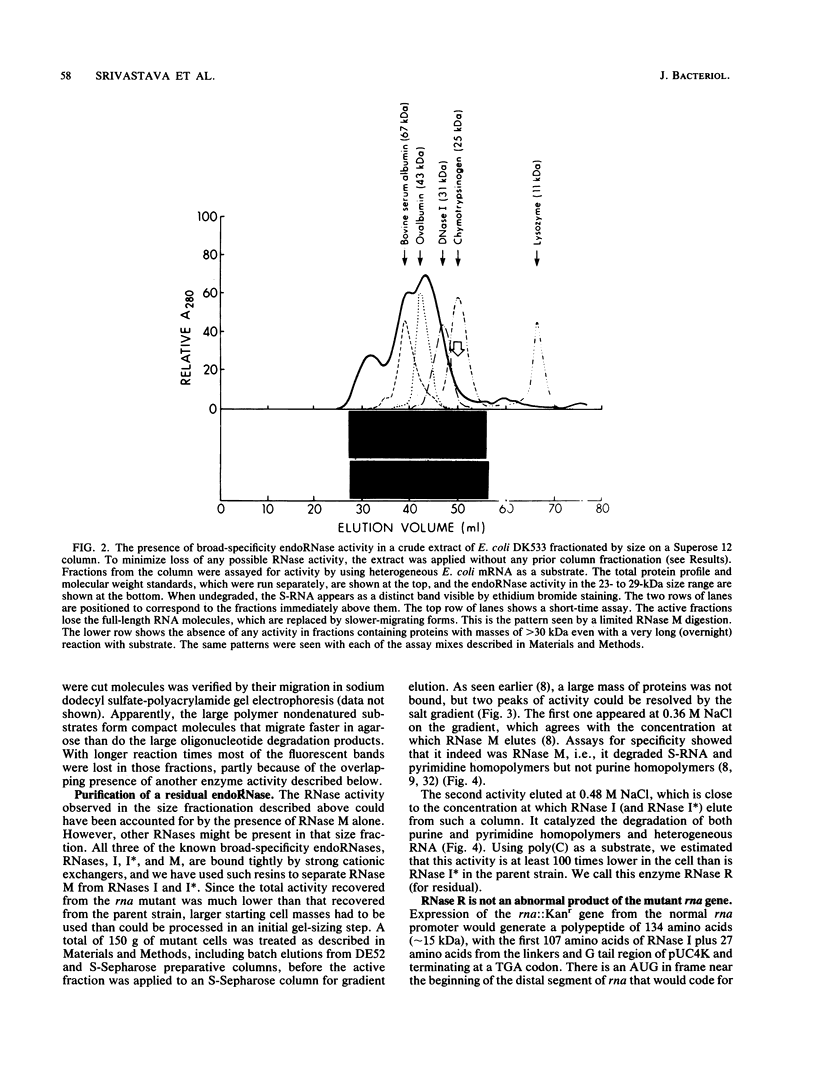
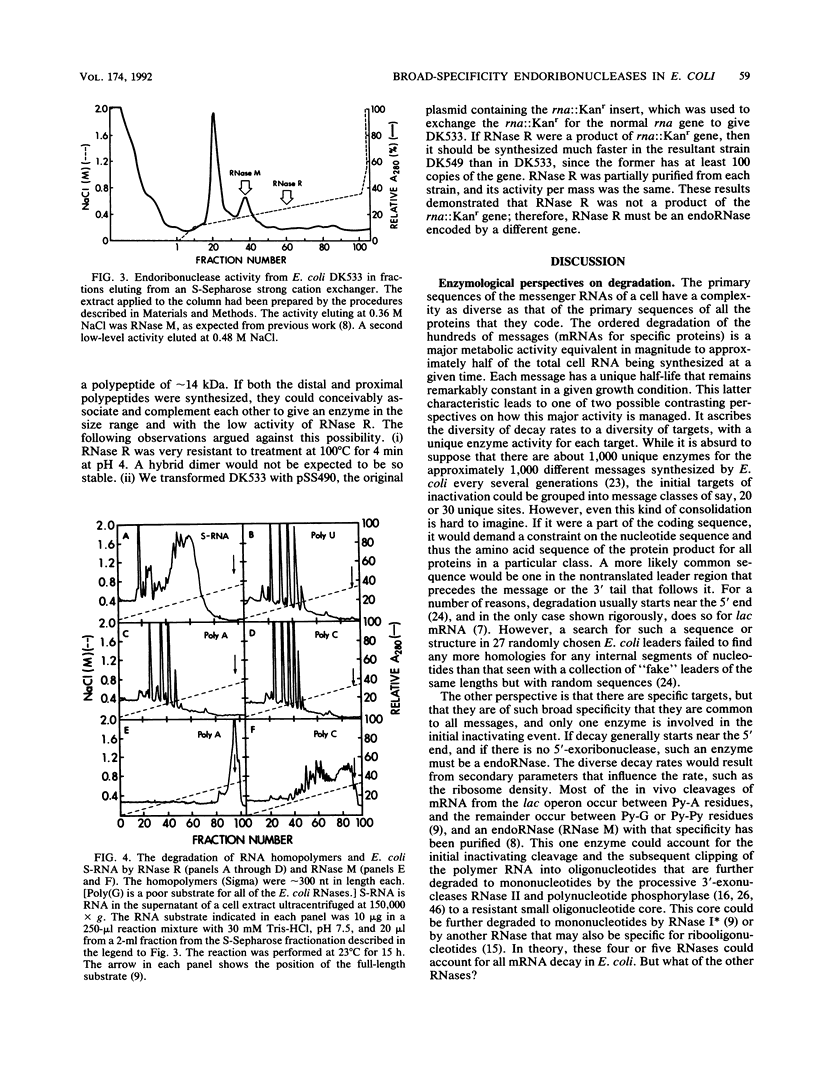



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Apirion D., Gitelman D. R. Decay of RNA in RNA processing mutants of Escherichia coli. Mol Gen Genet. 1980 Jan;177(2):339–343. doi: 10.1007/BF00267448. [DOI] [PubMed] [Google Scholar]
- Apirion D., Neil J., Watson N. Consequences of losing ribonuclease III on the Escherichia coli cell. Mol Gen Genet. 1976 Mar 22;144(2):185–190. doi: 10.1007/BF02428107. [DOI] [PubMed] [Google Scholar]
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Kushner S. R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett B. O., Scott Russell R., Jenkins W. Improved relationship between the deposition of strontium-90 and the contamination of milk in the United Kingdom. Nature. 1972 Jul 7;238(5358):46–48. doi: 10.1038/238046a0. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Evidence that the 5' end of lac mRNA starts to decay as soon as it is synthesized. J Bacteriol. 1985 Feb;161(2):820–822. doi: 10.1128/jb.161.2.820-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Purification and characterization of ribonuclease M and mRNA degradation in Escherichia coli. Eur J Biochem. 1989 May 1;181(2):363–370. doi: 10.1111/j.1432-1033.1989.tb14733.x. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli. J Bacteriol. 1991 Aug;173(15):4653–4659. doi: 10.1128/jb.173.15.4653-4659.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Wice B. M., Kennell D. E. Isolating and sequencing the predominant 5'-ends of a specific mRNA in cells. II. End-labeling and sequencing. J Biochem Biophys Methods. 1985 Aug;11(2-3):163–175. doi: 10.1016/0165-022x(85)90052-1. [DOI] [PubMed] [Google Scholar]
- Chanda P. K., Ono M., Kuwano M., Kung H. Cloning, sequence analysis, and expression of alteration of the mRNA stability gene (ams+) of Escherichia coli. J Bacteriol. 1985 Jan;161(1):446–449. doi: 10.1128/jb.161.1.446-449.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A. K., Miczak A., Taraseviciene L., Apirion D. Sequencing and expression of the rne gene of Escherichia coli. Nucleic Acids Res. 1991 Jan 11;19(1):125–129. doi: 10.1093/nar/19.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie-Martin F., Diaz-Torres M. R., Yancey S. D., Kushner S. R. Cloning of the altered mRNA stability (ams) gene of Escherichia coli K-12. J Bacteriol. 1989 Oct;171(10):5479–5486. doi: 10.1128/jb.171.10.5479-5486.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A. K., Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975 Sep 25;250(18):7313–7319. [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease 3. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GROS F., HIATT H., GILBERT W., KURLAND C. G., RISEBROUGH R. W., WATSON J. D. Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature. 1961 May 13;190:581–585. doi: 10.1038/190581a0. [DOI] [PubMed] [Google Scholar]
- Gottesman M., Oppenheim A., Court D. Retroregulation: control of gene expression from sites distal to the gene. Cell. 1982 Jul;29(3):727–728. doi: 10.1016/0092-8674(82)90434-2. [DOI] [PubMed] [Google Scholar]
- Hamilton C. M., Aldea M., Washburn B. K., Babitzke P., Kushner S. R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989 Sep;171(9):4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen S. M., Woolford C., van der Vies S. M., Tilly K., Dennis D. T., Georgopoulos C. P., Hendrix R. W., Ellis R. J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988 May 26;333(6171):330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Kennel D. Titration of the gene sites on DNA by DNA-RNA hybridization. II. The Escherichia coli chromosome. J Mol Biol. 1968 May 28;34(1):85–103. doi: 10.1016/0022-2836(68)90236-2. [DOI] [PubMed] [Google Scholar]
- Kennell D., Bicknell I. Decay of messenger ribonucleic acid from the lactose operon of Escherichia coli as a function of growth temperature. J Mol Biol. 1973 Feb 15;74(1):21–31. doi: 10.1016/0022-2836(73)90351-3. [DOI] [PubMed] [Google Scholar]
- Kinscherf T. G., Apirion D. Polynucleotide phosphorylase can participate in decay of mRNA in Escherichia coli in the absence of ribonuclease II. Mol Gen Genet. 1975 Sep 8;139(4):357–362. doi: 10.1007/BF00267975. [DOI] [PubMed] [Google Scholar]
- Klee C. B., Singer M. F. The processive degradation of individual polyribonucleotide chains. II. Micrococcus lysodeikticus polynucleotide phosphorylase. J Biol Chem. 1968 Mar 10;243(5):923–927. [PubMed] [Google Scholar]
- Kokoska R. J., Blumer K. J., Steege D. A. Phage fl mRNA processing in Escherichia coli: search for the upstream products of endonuclease cleavage, requirement for the product of the altered mRNA stability (ams) locus. Biochimie. 1990 Nov;72(11):803–811. doi: 10.1016/0300-9084(90)90189-n. [DOI] [PubMed] [Google Scholar]
- Lundberg U., von Gabain A., Melefors O. Cleavages in the 5' region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 1990 Sep;9(9):2731–2741. doi: 10.1002/j.1460-2075.1990.tb07460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie G. A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J Bacteriol. 1991 Apr;173(8):2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador J., 3rd, Cannon B., Cannistraro V. J., Kennell D. Purification and characterization of Escherichia coli RNase I. Comparisons with RNase M. Eur J Biochem. 1990 Feb 14;187(3):549–553. doi: 10.1111/j.1432-1033.1990.tb15336.x. [DOI] [PubMed] [Google Scholar]
- Meador J., 3rd, Kennell D. Cloning and sequencing the gene encoding Escherichia coli ribonuclease I: exact physical mapping using the genome library. Gene. 1990 Oct 30;95(1):1–7. doi: 10.1016/0378-1119(90)90406-h. [DOI] [PubMed] [Google Scholar]
- Misra T. K., Apirion D. RNase E, an RNA processing enzyme from Escherichia coli. J Biol Chem. 1979 Nov 10;254(21):11154–11159. [PubMed] [Google Scholar]
- Mudd E. A., Krisch H. M., Higgins C. F. RNase E, an endoribonuclease, has a general role in the chemical decay of Escherichia coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990 Dec;4(12):2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Silengo L., Schlessinger D. Synthesis of a large precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M., Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of messenger RNA. J Mol Biol. 1979 Apr 15;129(3):343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- Ono M., Kuwano M. Chromosomal location of a gene for chemical longevity of messenger ribonculeic acid in a temperature-sensitive mutant of Escherichia coli. J Bacteriol. 1980 Apr;142(1):325–326. doi: 10.1128/jb.142.1.325-326.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. Functional mRNA half lives in E. coli. Mol Gen Genet. 1978 Nov 9;166(3):329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. K., Singh B., Ray B. K., Apirion D. Maturation of 5-S rRNA: ribonuclease E cleavages and their dependence on precursor sequences. Eur J Biochem. 1983 Mar 1;131(1):119–127. doi: 10.1111/j.1432-1033.1983.tb07238.x. [DOI] [PubMed] [Google Scholar]
- Schindler D., Echols H. Retroregulation of the int gene of bacteriophage lambda: control of translation completion. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4475–4479. doi: 10.1073/pnas.78.7.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Singer M. F., Tolbert G. Purification and properties of a potassium-activated phosphodiesterase (RNAase II) from Escherichia coli. Biochemistry. 1965 Jul;4(7):1319–1330. doi: 10.1021/bi00883a016. [DOI] [PubMed] [Google Scholar]
- Talkad V., Achord D., Kennell D. Altered mRNA metabolism in ribonuclease III-deficient strains of Escherichia coli. J Bacteriol. 1978 Aug;135(2):528–541. doi: 10.1128/jb.135.2.528-541.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraseviciene L., Miczak A., Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991 Apr;5(4):851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- Taylor L. A., Rose R. E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988 Jan 11;16(1):358–358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomcsányi T., Apirion D. Processing enzyme ribonuclease E specifically cleaves RNA I. An inhibitor of primer formation in plasmid DNA synthesis. J Mol Biol. 1985 Oct 20;185(4):713–720. doi: 10.1016/0022-2836(85)90056-7. [DOI] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]