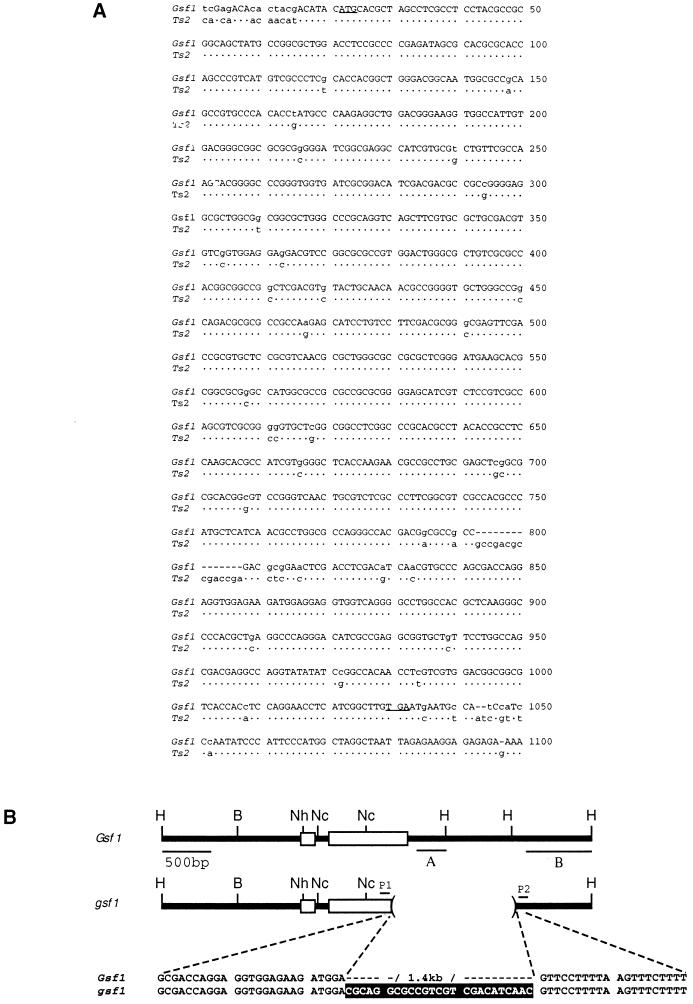
Figure 5.
Gsf1 and Ts2 genes. (A) Shown are the Gsf1 coding sequence (top line) with the start and stop codons underlined and the mismatches in the Ts2 gene sequence indicated below. The coding sequences from Gsf1 and Ts2 show more than 90% identity. (B) Results from Southern analysis and DNA sequencing of the gsf1 mutation are summarized. Probe A hybridized to Gsf1 DNA but not to gsf1 DNA. Probe B hybridized to both genomic DNAs, but the gsf1 fragment was 1.4 kb smaller than in wild type (data not shown). PCR analysis using P1 and P2 as primers also indicated that the gsf1 amplification product was 1.4 kb smaller than the product found in wild type. Sequence analysis of both alleles indicated that, between the deletion breakpoints, 25 bp of filler DNA is inserted (highlighted nucleotides). In addition to the 1.4-kb deletion, several minor nucleotide polymorphisms in intron and 5′ noncoding sequences were detected between wild-type and mutant alleles (not shown). H, HindIII; B, BamHI; Nh, NheI; Nc, NcoI.