Abstract
Over the past few years, technological advances in automated DNA sequencing have had a profound effect on the nature of DNA sequencing laboratories. To characterize the changes occurring within DNA sequencing facilities, the DNA Sequencing Research Group conducted three previous studies, in 1998, 2000, and 2003. A new general survey has been designed and conducted by the DSRG to capture the current status of DNA sequencing facilities in all sectors. Included were questions regarding facility administration, pricing, instrumentation, technology, protocols, and operation. The results of the survey are presented here, accompanied by comparisons to the previous surveys. These comparisons formed a basis for the discussion of trends within the facilities in response to the dynamics of a changing technology.
Keywords: Automated DNA sequencing, survey, DNA sequencing facility, DNA sequencing chemistry
Since 1998, the DNA Sequencing Research Group (DSRG) has conducted three general surveys (1998, 2000, and 2003) designed to collect data on the makeup and operation of DNA sequencing facilities. For the 2006 study, the DSRG prepared a more comprehensive survey consisting of ninety questions, which were divided into five major categories: (1) the makeup of the facility, (2) services and support, (3) pricing, (4) instrumentation, and (5) sequencing reactions.
METHODOLOGY
The survey was launched on November 10, 2005. In an effort to ensure wide distribution, the survey notification was posted on both the Association of Biomolecular Resource Facilities (ABRF.org) Web site and listserv, as well as the Evolutionary Directory (http://evol.mcmaster.ca/brian/evoldir.html) and the SIOSCI/Bionet Automated Sequencing (www.bio.net/biomail/listinfo/auto-seq) newsgroups. All submissions were accepted until the closing date of December 31, 2005, with a request that a manager or director submit only one response from a given facility. Responses were received from 61 laboratories around the world. Of the respondents, 38 were from the United States, 10 from Canada, 8 from Europe, and 1 each from Australia, Japan, India, Saudi Arabia, and South Africa. The majority of respondents (74%) were members of ABRF. Preliminary results of the survey were collected, analyzed, and presented at the 2006 annual ABRF conference.1,2 Here, we present a more detailed report of the survey data. To facilitate comparisons between countries, all monetary data have been converted to US currency.
RESULTS AND DISCUSSION
Facility Makeup
The first portion of the questionnaire, facility makeup, focused on composition, funding, and administration. Information was also gathered regarding personnel, including educational backgrounds and salaries. We expected that these questions would provide us with an overview as well as the baseline for all operational aspects of current DNA sequencing facilities.
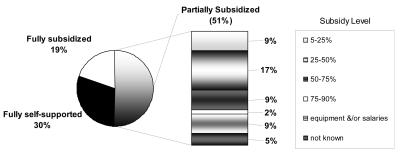
The majority of the survey participants (43) indicated an academic affiliation. Of the remaining respondents, 6 operated as commercial or fee-for-service facilities, 5 were government laboratories, 4 reported being private research organizations, and 3 were hospital laboratories. The majority of laboratories, 44 out of 61, had been providing DNA sequencing services for five years or longer. Only 2 of the laboratories had been operating for less than one year. Most facilities responded that they offered additional services, such as fragment analysis (74%), DNA preparation (38%), SNP discovery and confirmation (31%), microarray (4%), and RT/quantitative-PCR (6%). A small percentage (11%) of the laboratories indicated capability to provide other services, such as library construction, colony picking, DNA synthesis, proteomics, phosphor-imaging, or reagent stores. When asked about the funding of the facility, only 30% responded that they were fully self-supported. Figure 1 provides an overview of reported level of subsidy of the partially subsidized facilities.
FIGURE 1.
Facility funding.
A vast majority of the laboratories (79%) responded that the facility was governed by either an external faculty advisor or by an advisory committee. Within the facility, the personnel typically consisted of a director or manager, and three technicians. In comparison, the 2003 DSRG sequencing survey reported that the average laboratory consisted of five individuals: a director or manager, and four technical staff.3 The decrease in full-time staff most likely reflects the increased percent of facilities operating high-throughput capillary platforms, as is discussed in a later section. A slim majority (55%) of the participants reported that the directors or managers held PhD and/or MBA degrees. Of the remaining directors or managers, 20% reported having a master’s degree and 25% held bachelor’s degrees. The majority (62%) of the technical staff within the facility held bachelor’s degrees, although 38% indicated advanced degree levels.
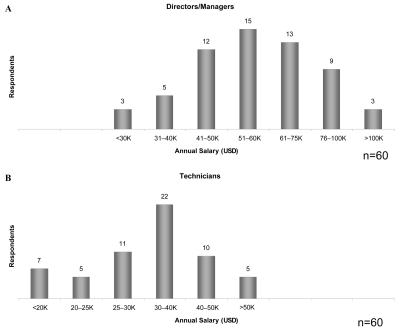
The salaries reported for the directors/managers were found to be distributed over a wide range, from under $30,000 to over $100,000 per annum, with the average annual salary ranging between $51,000 and $60,000 (Figure 2A). Similarly, the technical staff salaries were also distributed over a wide range, from under $20,000 to over $50,000 per annum, with a median annual salary range of $30,000 to $40,000 (Figure 2B). In the 2003 survey, the average salary for the director was reported to be $58,000 and the average salary for the full-time staff was $32,000. It seems that the compensation of both managers and technical staff has remained largely unchanged since 2003.
FIGURE 2.
Annual salary ranges, in us dollars, for (A) directors/managers and (B) technicians.
Most of the DNA sequencing facilities participating in this survey have been operational for several years and are located within academic institutions. They are maintained by a small, professionally educated group of staff, administered by either a director or a manager. In addition to providing DNA sequencing services, most of the facilities are engaged in offering a variety of additional biotechnical support to the research community.
Services and Support
This section of the questionnaire was designed to elucidate the operational functions of facilities, such as the number of sequencing reactions processed within the laboratory, evaluation of the reaction performance, turnaround time, additional services offered, and the manner of data delivery.
The submission and processing of sequencing work orders greatly varied from one facility to another. In general, facilities allow investigators flexibility in the submission of sequencing requests. The preferred method was submission through a LIMS (43%), although many facilities indicated acceptance of both printed and electronic forms (27%). Types of electronic submissions included electronic spreadsheets or email requests. A small number of respondents (13%) indicated that they accepted only paper submissions, whereas some facilities (7%) reported accepting printed submissions, which were subsequently entered into a LIMS.
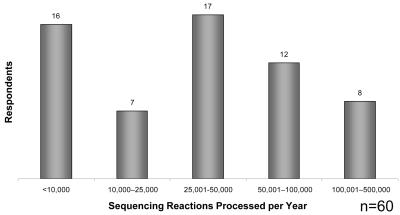
The number of sequencing reactions completed within the previous twelve months varied widely from facility to facility, from less than 10,000 to as many as 500,000 reactions per year (Figure 3). The average laboratory completed between 25,000 and 50,000 reactions. Overall, these numbers have significantly increased, based on responses from the 19984 and 20005 sequencing surveys. Even though the average number of sequencing reactions does not appear to have changed from the 2003 survey (Table 1), the largest reported number of sequencing reactions in 2003 was 250,000 and in 2006 was 500,000, suggesting that the upper limits seem to be shifting towards 500,000+ per annum.
FIGURE 3.
Number of processed sequencing reactions within the year.
TABLE 1.
Processed Sequences per Year
| Survey year | Average No. Samples | N |
|---|---|---|
| 1998 | 11,742 | 59 |
| 2000 | 18,813 | 37 |
| 2003 | 47,000 | 30 |
| 2006 | 25,000–50,000 | 61 |
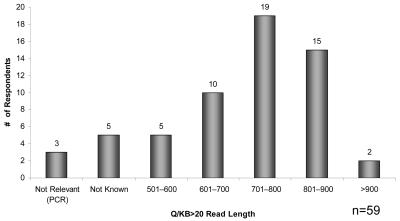
The participants were asked to assess the quality of their DNA sequence data in terms of Q > 20 length of read.6 Fifty-nine facilities responded to this question. These results are shown in Figure 4. Of these, three facilities reported that they sequenced very short PCR fragments, and considered the Q/KB > 20 values to be irrelevant. Five other facilities responded that this value was unknown, either due to the lack of a proper reporting software or irrelevance of such measures for their operations. All of the remaining fifty-one laboratories reported Q20 read lengths ranging from 501 bases to over 900 bases, with a median quality read length in the 701–800 base range.
FIGURE 4.
Assignment of sequence quality by the respondents.
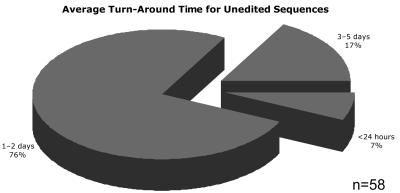
A question most often asked by facility users is, “How long will it be until I receive my data?” This question was also included in the survey, and it is illustrated in Figure 5. The reported turnaround time for processing unedited sequences varied from 4 h to 5 d, with an average of 24 to 48 h. In comparison, an average turnaround time of 48 h was reported in 1998. Of note is that 7% of the sequencing facilities currently reported an average turnaround time of less than 24 h.
FIGURE 5.
Percentage of respondents reporting average turnaround time for unedited sequences.
Pricing
The third portion of the survey dealt with pricing structure for work performed. Some core facilities reported using a multitiered pricing system comprised of one price for external projects and another price for in-house projects, with provisions for bulk discounts. Although data were collected for all situations, the comparisons presented are only for in-house pricing of a single sequencing reaction. Worldwide, the sequencing prices varied widely, from $1.35 to $30.00 per reaction. Within the United States, the maximum price for a single sequencing reaction was $15.00 (Table 2). When comparing these findings to those from previous surveys, the average price for a sequencing reaction had dropped significantly, from $14.92 as reported in the 1998 survey to under $9.00. Not surprisingly, the participating laboratories reported that 74% of their customers were satisfied with this pricing.
TABLE 2.
Price per Sequencing Reaction (US Dollars)
| Total Respondents | USA | Canada | Europe | Other | |
|---|---|---|---|---|---|
| N | 45 | 29 | 7 | 5 | 4 |
| Range | $1.35–$30.00 | $1.35–$15.00 | $5.27–$21.95 | $4.00–$8.61 | $12.00–$30.00 |
| Median | $8.78 | $9.00 | $8.78 | $6.07 | $15.09 |
| Mode | $10.00 | $10.00 | $8.78 | N/A | N/A |
Instrumentation
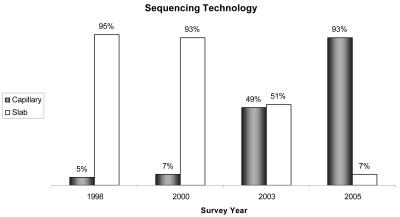
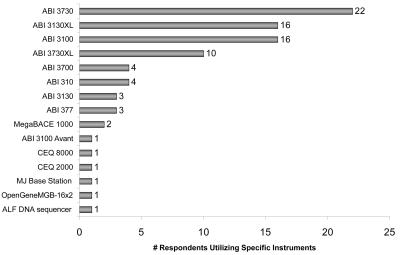
This section queried survey participants on the number and type of sequencing platforms owned by and operated within the facility. Figure 6 shows that in the past three years, capillary electrophoresis has become the dominating platform. An overwhelming majority (93%) of the participants report processing samples on capillary instruments in contrast to using a slab gel system (7%). Furthermore, 90% of the respondents reported having only capillary systems, while the remaining 10% were evenly divided between having only slab gels or having a combination of slab gels and capillary systems. In comparison, the 2003 study indicated that 40% of the core facilities had only capillary instruments, 37% had only slab gels, and 23% had both platforms. When asked which commercial sequencing platform was used within the facility, the majority of the participants responded, as they had in previous surveys, that they operated an Applied Biosystems sequencing platform (Figure 7).
FIGURE 6.
Comparison of DNA sequencing platforms.
FIGURE 7.
Type of sequencing platforms reported by respondents.
Sequencing Chemistry and Reaction Cleanup
The last section of the questionnaire requested detailed information on sequencing setups and protocols. Questions were asked to determine the type of reaction vessels employed, sequencing chemistries, and post-reaction cleanup methods used to remove unincorporated dye terminators.
The introduction of capillary electrophoretic systems, with increased sample-handling capabilities, has resulted in a corresponding change in the sample reaction vessels. In 2003, there was an even distribution of facilities performing reactions in single tubes, 96-well plates, or a combination of both. Currently, 72% of facilities reported strictly using plate format (384-well, 96-well, or 8 × 12 strip tubes), 7% reported using single-tube format, and 21% reported a combination of plate and tube formats. Two respondents indicated using 384-well plates exclusively.
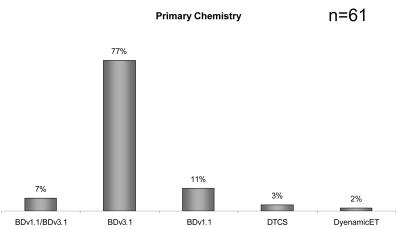
To determine the type of sequencing chemistry utilized within each facility, respondents were asked to indicate their primary chemistry method, all additional chemistries, as well as reaction additives employed. The results are presented in Figure 8. A vast majority (77%) identified BigDye Terminator v3.1 Chemistry (Applied Biosystems) as their primary chemistry of choice. The second most popular primary chemistry was BigDye Terminator 1.1 chemistry (11%), and 7% of respondents reported alternating between BigDye Terminator v1.1 and v3.1. The remaining respondents used either the Dye Terminator Cycle Sequencing (DTCS) kit (3%) for sequencing on CEQ Genetic Analysis Systems from Beckman Coulter, Inc., or DYEnamic ET Dye Terminator Kit (2%) for sequencing on MegaBACE DNA Analysis Systems (GE Healthcare Life Sciences). These responses appear to reflect the type of commercial sequencing platform the facilities reported. In the 2003 survey, BigDye Terminator chemistry was also reported to be the dominant method of choice. For respondents indicating a secondary sequencing chemistry, 28% used ABI PRISM dGTP BigDye Terminator v3.0 chemistry, and 12% used ABI PRISM BigDye Terminator 1.1 chemistry. In 71% of the laboratories, additives were regularly used to sequence through difficult regions. The two most common additives reported were DMSO (41%) and betaine (38%).
FIGURE 8.
Primary DNA sequencing chemistry.
Many diverse methods are available for removing unincorporated dye terminators and primers prior to sample loading onto sequencing instruments. The respondents were equally divided between using commercial 96-well filter plates (32%) or EtOH precipitation (31%). Some facilities (15%) also prepared their own 96-well filter plates. The use of magnetic beads constituted 15% of the responses. Only 7% reported using individual size exclusion (chromatography) columns. In comparison, the 2003 survey revealed 62% of respondents utilizing size-exclusion columns.
SUMMARY
Over the past six years, the DSRG has conducted three general surveys of DNA sequencing facilities. Based on the responses by participating laboratories, several trends can be noted: (1) The average number of personnel within the facility appears to have decreased (from five to four); (2) there is a wide distribution of salaries for both the directors/managers and staff; (3) the majority of the laboratories are subsidized; (4) the number of sequencing reactions performed by the facilities has increased from past years, but may be leveling; (5) the average sequencing chargeback has significantly decreased (by approximately 40%); (6) there has been a significant change from gel-based platforms to capillary-based platforms from the survey conducted in 2003; (7) the Applied Biosystems instruments are the dominant capillary platform (92%); (8)BigDye Terminator v3.1 Chemistry is the dominant chemistry, and the use of additives is common when sequencing difficult templates; and (9) size-exclusion or chromatography columns are no longer the preferred method for cleanup of sequencing reactions.
The change from gel-based platforms to capillary-based platforms has been a driving force in determining the profile of DNA sequencing facilities. With the advent of new technologies that are addressing the rapid, low-cost sequencing of genomes, we envision that in the coming years these facilities will be different in nature and scope. On the other hand, we also believe that the current capillary technology (or some modification of it) will be in use in core facilities for many years to come. We hope that this comprehensive survey not only provides a measuring stick and reference for any individual sequencing core facility, but will also serve as a basis for comparison for future surveys.
ACKNOWLEDGMENTS
The members of the DSRG wish to thank everyone who participated in the survey as well as Chris Walls (University of Utah) for implementation of the survey design. Our special thanks to Susan Hardin and to Pamela S. Adams, who have respectively served as our past and present liaisons between the DSRG and the ABRF Executive Board. Mention of trade names, commercial instruments, or commercial products does not constitute an endorsement or recommendation for use.
REFERENCES
- 1.Wiebe GJ, Schweitzer PA, Needleman DS, Adam D, Detwiler M, Escobar H, et al. DSRG Presentation: 2006 General Survey of DNA Sequencing Facilities and Launch of the Web-based Trouble-Shooting Resource. J Biomol Tech. 2006;17:73. (Abstr) [PMC free article] [PubMed] [Google Scholar]
- 2.Adam D, Kieleczawa J, Escobar H, Detwiler M, Pershad R, Needleman DS, et al. DNA Sequencing Research Group—2006 General Survey of DNA Sequencing Facilities. J Biomol Tech. 2006;17:28. (Abstr) [PMC free article] [PubMed] [Google Scholar]
- 3.Wiebe GJ, Pershad R, Escobar H, Hawes JW, Hunter T, Jackson-Machelski E, et al. DNA Sequencing Research Group (DSRG) 2003—A General Survey of Core DNA Sequencing Facilities. J Biomol Tech. 2003;14:231–237. [Google Scholar]
- 4.Grills G, Adams PS, Dolejsi MK, Hardin S, McMinimy D, Morrison P, et al. Analysis of the Effects of Different DNA Sequencing Methods on Sequencing Quality, Creation of a Quality Control Resource, and Assessment of the Current State of the Art: Results from the 1998 ABRF DNA Sequence Research Committee Study. http://www.abrf.org/ResearchGroups/DNASequencing/EPosters/dsrg98/survey/Fig9.htm.
- 5.Leviten D, Grills G, Hardin S, Robertson M, Thannhauser T, VanEe J. Results from the DNA Sequencing Research Group’s Survey of Core Facilities, 2000. ABRF DSRG 2000 Web Poster. www.abrf.org/ResearchGroups/DNASequencing/EPosters/DSRGGeneral2k/index.htm.
- 6.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]