Figure 1. Map of the P. murina expression-site locus.
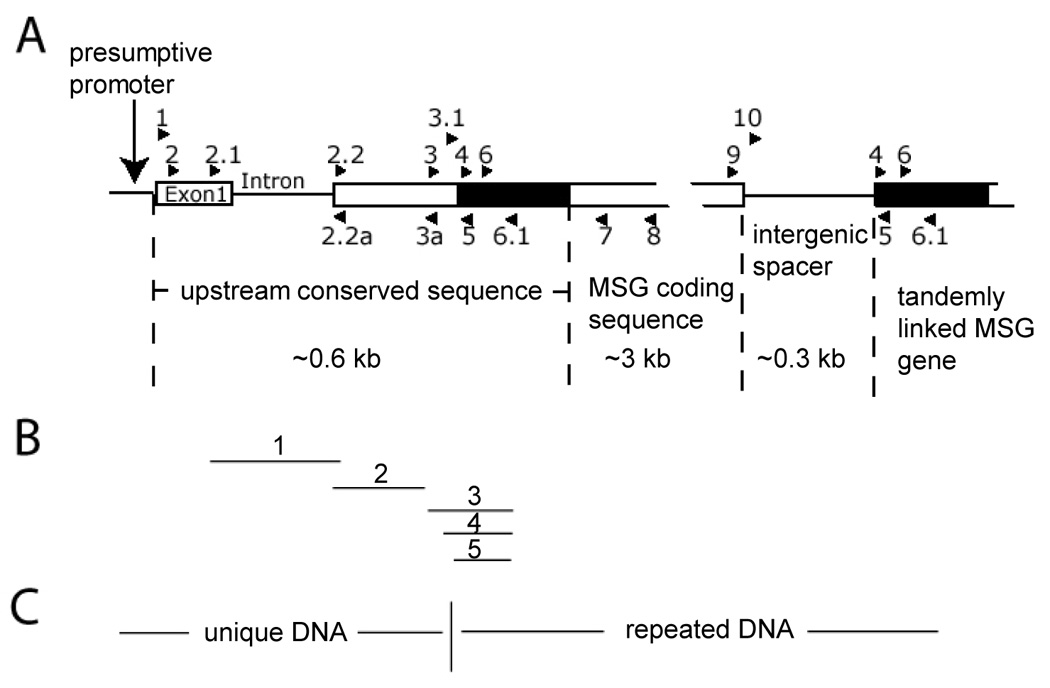
A) The map was constructed primarily from information obtained in the studies presented herein. The map begins upstream of the sequence analyzed. Presumably, there is a promoter of transcription located in this region. Exon 1 marks the beginning of the sequence that encodes the Upstream Conserved Sequence (UCS), which is the sequence found at the beginning of messenger RNAs encoding diverse MSGs. DNA encoding the UCS extends from exon 1, which is unique in the genome, through the intron into a second block of unique coding DNA. This block of unique DNA (exon 1-intron-beginning of exon 2) constitutes the expression site. Adjacent to the expression site is a block of non-unique DNA called the CRJE (see Table 1), which is represented by a black rectangle. The 3-prime end of the UCS coincides with the 3-prime end of the CRJE. Other copies of the CRJE are located at the beginning of MSG genes not at the expression site. Hence the beginning of the UCS is encoded by unique DNA, and the end is encoded repeated DNA. The sequence encoding the UCS is followed by a sequence encoding an MSG protein. Two tandem MSG-encoding sequences are shown. The first is directly attached to the expression site; the second lies downstream of the first, and is separated from the UCS-linked MSG gene by approximately 300 bp (intergenic spacer). It should be noted that while tandem gene pairs are demonstrated by the data presented herein, it is not yet known if a tandem gene pair exists at the expression site. Locations and names of PCR primers are indicated by numbered arrowheads. See Table 2 for primer sequences and locations. B) The five numbered lines mark regions studied by real-time PCR. C) The region to the left of the line contains a DNA sequence that is present only once in the haploid genome of P. murina. The region to the right of the line contains a DNA sequence that is present in multiple copies in the genome.
