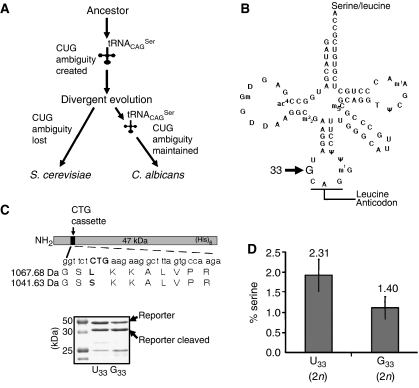
Figure 1.
Reconstruction model of the Candida genetic code alteration. (A) Redefinition of the identity of the CUG codon from leucine to serine in Candida started with a novel serine tRNA (tRNACAGSer) and evolved gradually over the last 272±25 million years. tRNACAGSer disappeared and the cognate leucine CUG decoder (tRNACAGLeu) was maintained in the S. cerevisiae lineage (standard genetic code), while the converse occurred in the C. albicans lineage (altered genetic code). (B) The tRNACAGSer contains guanosine at position 33 (G33), which is a conserved position occupied by uridine (U33; U-turn) in other tRNAs. (C) The upper panel shows a diagram of the reporter system used to quantify serine misincorporation at CUG codons in vivo in S. cerevisiae. A CUG cassette inserted in the CaPGK gene was flanked by two thrombin cleavage sites to facilitate the purification of the short reporter peptide encoded by the cassette. The recombinant protein was expressed and purified from S. cerevisiae cultures using nickel affinity chromatography, and was then cleaved with thrombin for 16 h at 26°C, in solution. The resulting peptides were analysed by mass spectrometry. The lower panel shows a 12% SDS–PAGE of the reporter protein. (D) Serine and leucine incorporation at the CUG position (see panel C) was determined by quantitative MRM methodologies using a hybrid quadrupole/linear ion-trap mass spectrometer. Synthetic peptides with sequences identical to those of the serine and leucine peptides shown in panel C were used as external controls and to build the calibration curves used for quantification.