Figure 1.
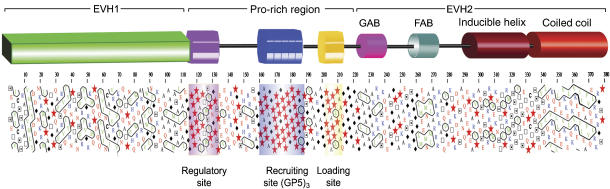
Modular organization of VASP. The diagram shown on the upper part of the figure represents the modular organization of the human VASP sequence (UniProt P50552) based on the distribution of clusters of hydrophobic amino acids determined with the program HCA (Callebaut et al, 1997). Symbols are as follows: Pro,  ; Gly, ⧫; Thr, □ and Ser,
; Gly, ⧫; Thr, □ and Ser,  . Amino acids are colored according to their chemical characteristics: green, hydrophobic; red, negatively charged; blue, positively charged; black, Ala and Cys. The EVH1 and CC domains, which are stably folded and whose structures have been determined (Prehoda et al, 1999; Kuhnel et al, 2004), are characterized by a higher density of clusters of hydrophobic amino acids (contoured black). In contrast, the extended Pro-rich region (amino acids 116–213) lacks hydrophobic clusters, and is predicted to be mostly unfolded. Within this region, we identify three distinct groups of Pro residues, which are present in all members of the Ena/VASP family: regulatory site (116–135), recruiting site (160–194) and loading site (201–211). Finally, the region preceding the CC domain has the typical HCA pattern of an inducible α-helix.
. Amino acids are colored according to their chemical characteristics: green, hydrophobic; red, negatively charged; blue, positively charged; black, Ala and Cys. The EVH1 and CC domains, which are stably folded and whose structures have been determined (Prehoda et al, 1999; Kuhnel et al, 2004), are characterized by a higher density of clusters of hydrophobic amino acids (contoured black). In contrast, the extended Pro-rich region (amino acids 116–213) lacks hydrophobic clusters, and is predicted to be mostly unfolded. Within this region, we identify three distinct groups of Pro residues, which are present in all members of the Ena/VASP family: regulatory site (116–135), recruiting site (160–194) and loading site (201–211). Finally, the region preceding the CC domain has the typical HCA pattern of an inducible α-helix.