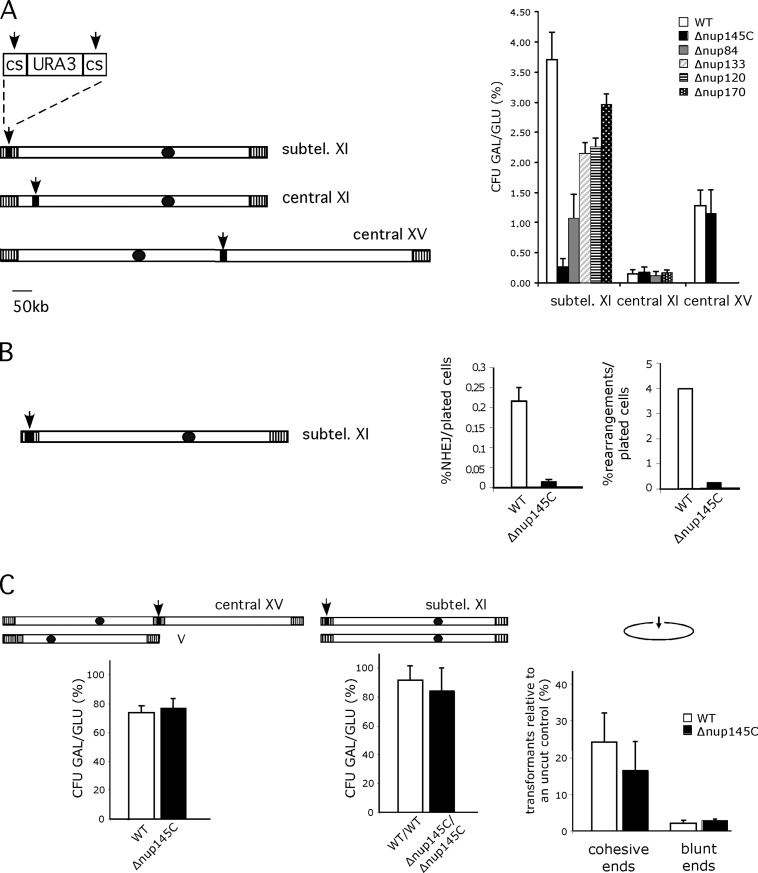
Figure 2.
Nup84 complex is required for subtelomeric DSB repair efficiencies. (A) Survival to DSBs according to their position in wild-type and different nucleoporin mutant backgrounds. Chromosomes XI and XV are schematized. Black dots and vertical hatched boxes represent centromere and subtelomeres, respectively. Arrows indicate the positions of induced DSBs generated by cleavage of two inverted I-SceI cutting sites (cs) surrounding the URA3 marker (black box). Size in kb is indicated. Colony forming units (CFU) are calculated as the ratio of galactose growing colonies (GAL) versus glucose growing colonies (GLU) expressed in percentages (%). Values obtained for wild-type (WT, white), Δnup145C (black), Δnup84 (gray), Δnup133 (diagonals), Δnup120 (horizontal lines), and Δnup170 (white dots) are shown according to DSB's position. Subtel. XI stands for subtelomeric XI-L. Standard errors with a P value of 0.1 are shown. (B) Proportion of repair by NHEJ and rearrangements in subtelomere XI-L. Subtelomeric DSB is schematized as in A. Percentages of repair either by NHEJ or by gross chromosomal rearrangements per plated cells are shown for wild-type (WT, white) and Δnup145C (black) (C). Repair machinery is functional in Δnup145C. Left; DSB is generated in a central part of chromosome XV and gene conversion is made possible because of homologous sequences present on chromosome V (gray box, see text and Karathanasis and Wilson, 2002). Colony forming units are calculated as in A. Middle; DSB is generated in the subtelomeric position in diploid wild-type cells or diploid cells homozygous for Δnup145C. Colony forming units are calculated as in A. Right; DSB is created on a replicative plasmid by restriction enzymes generating cohesive or blunt ended ends. Wild-type (WT) and Δnup145C strains were transformed with equivalent amounts of supercoiled or linearized pRS316 (ARS-CEN-URA3) plasmid. The number of transformants obtained with the linearized plasmid expressed as a percentage of the number of transformants obtained with the supercoil DNA are plotted. Experiments were repeated three times and error bars represent the 95% confidence intervals.