Figure 1. Identification of a Slimb binding site on Ci between PKA sites 1 and 2.
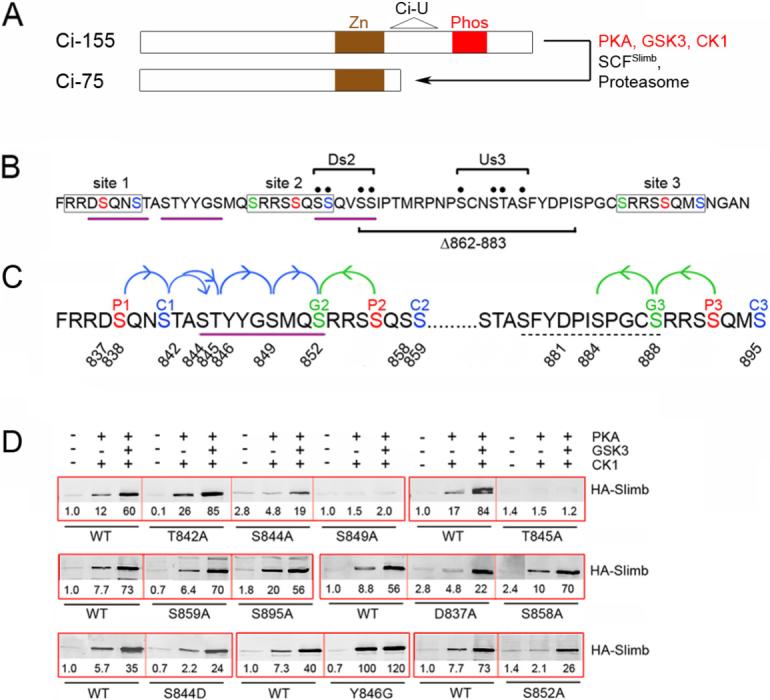
(A) Schematic showing the Zinc finger (Zn) DNA-binding domain shared by Ci-155 and Ci-75, the phosphorylated region (Phos) depicted in B and C, and the region of Ci deleted in the Ci-U transgene. (B) Amino acids 834−899 of Ci showing the three (lightly boxed) groups of PKA sites (red, P1-P3), PKA-primed GSK3 sites (green, G2 and G3) and PKA-primed CK1 sites (blue, C1-C3). Motifs resembling a consensus (DSGxxS) Slimb binding site are underlined in purple. The four Ala substitutions of Ci-Ds2 and Ci-Us3 (bracketed black dots) and residues deleted in Ci-Δ862−883 are indicated. (C) Single Ala substituents tested in this study are indicated by residue number. The extended Slimb binding motif defined here is underlined (purple) and arrows indicate critical consecutive CK1 (blue) and GSK3 (green) phosphorylations. Dashed underlining indicates residues of a potential weak Slimb binding motif. (D) GST-Ci proteins with the named substitutions were phosphorylated by the indicated protein kinases and incubated with extracts of Kc cells transfected with an HA/Flag-Slimb-Myc expression vector. Proteins brought down with glutathione beads were visualized on Western blots with HA antibody. In this and other Figures, thick red boxes group single experiments that test GST-Ci variants relative to each other and a wild-type (WT) control strictly in parallel. HA-Slimb band intensities were measured, corrected for background and expressed relative to GST-Ci-WT with no phosphorylation (fixed at 1.0) for that experiment. GST blots (not shown) confirm similar protein levels and similarly efficient phosphorylation, judged by mobility shifts.
