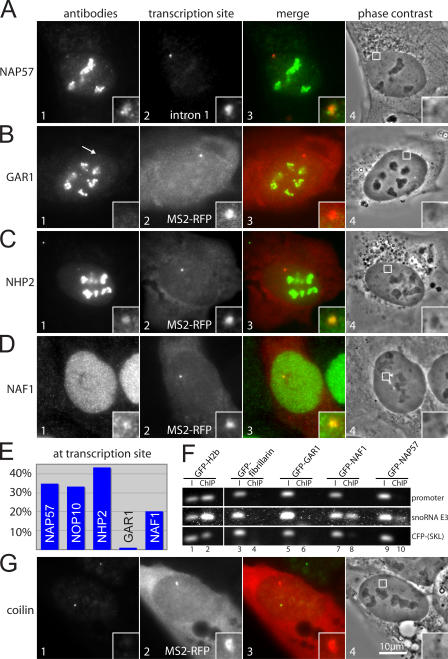
Figure 5.
Colocalization of endogenous proteins with the H/ACA RNA transcription site. (A) Double fluorescence of a fixed and permeabilized E3 cell after induction of the transgene and immunostained for endogenous NAP57 (panel 1) and for the H/ACA RNA transcription site by FISH with the probe to intron 1 (panel 2). Panel 3 shows a merge of the two in pseudocolor. Panel 4 depicts the corresponding phase-contrast image outlining, with a box indicating the area containing the transcription site, which is enlarged in the insets. (B–D) Same as in A, except immunostained for the endogenous proteins indicated on the left, and the transcription site was identified by the fluorescence of transfected MS2-RFP (panel 2). (E) Bar diagram of the percentage of cells, with the indicated proteins observed at the site of H/ACA RNA transcription. The following numbers of cells with a transcription site (in parentheses) were counted for each protein: NAP57 (150); NOP10 (101); NHP2 (101); GAR1 (104); and NAF1 (159). Questionable colocalizations were discounted. (F) ChIPs of the GFP-fusion constructs indicated on top after their transient transfection into E3 cells and induction of the transgene. Immunoprecipitations were performed with GFP antibodies. Ethidium bromide stain of DNA amplified by PCR, with primers to the regions of the transgene indicated on the right and separated on agarose gels. Amplifications of the input (I; odd lanes) and the ChIPs (even lanes) are shown. (G) Same as in B and C, except stained for the Cajal body marker protein coilin.