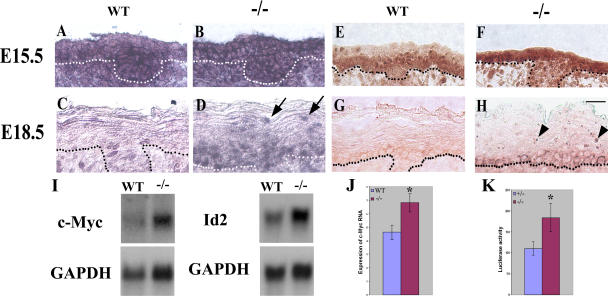
Figure 5.
c-myc expression is up-regulated in developing suprabasal layers of the Ovol1-deficient epidermis. Results of situ hybridization (A–D) and immunostaining (E–H) of wild-type (A, C, E, and G) and mutant (B, D, F, and H) embryonic skin using a c-myc cRNA probe and a mouse anti–c-myc antibody, respectively. Note that at E18.5, c-myc protein is cytoplasmic in the basal layer, and at least some c-myc transcripts appeared nuclear, which probably stems from the reported delicate control of subcellular localizations of c-myc gene products to tailor the needs of cell cycle control in these cells (Vriz et al., 1992; Bond and Wold, 1993). Dotted lines denote the basement membrane. Arrows and arrowheads indicate mutant suprabasal cells expressing c-myc mRNA and protein, respectively. (I) Results of Northern blot analysis of E16.5 wild-type and mutant skin. Each blot was stripped and reprobed with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe. (J and K) Real-time PCR analysis of c-myc transcript levels (J) and reporter assays of c-myc promoter activity (K) in wild-type and mutant keratinocytes 24 h after Ca2+ induction. Average values obtained on cells from three mutant and three control animals are shown. Luciferase activities were normalized for transfection efficiency by using a β-actin promoter driving lacZ as an internal control. *, P < 0.03 difference from the wild type. Error bars represent SEM. Bar, 20 μm.