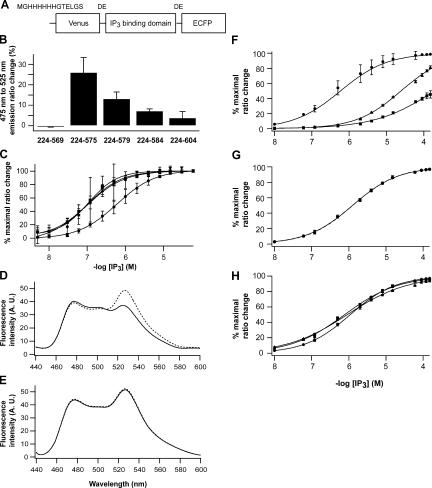
Figure 1.
IP3 sensor proteins based on the IP3 binding domain of mouse IP3R1. (A) A basic design for IP3 sensors. (B) ECFP/Venus emission ratio changes of IP3 sensors in COS7 lysates after the addition of 60 μM of IP3. (C) Apparent IP3 affinity of IP3 sensors composed with amino acid residues 224–575 (circle), 224–579 (triangle), 224–584 (square), and 224–604 (inverse triangle) in COS7 lysates. Data were obtained from three independent measurements. Emission spectra of purified IRIS-1 (D) and IRIS-1–Dmut (E) excited at 420 nm with 0 μM (broken line) and 100 μM (solid line) of IP3. Measurements were performed at 20°C in buffer A (10 mM Hepes, pH 7.2, 100 mM NaCl, 1 mM 2-mercaptoethanol, and 0.5% NP-40) containing 1 mM EDTA. (F) Specificity of IRIS-1 against IP3 (circle) and its natural metabolites, 1,3,4,5 IP4 (triangle) and 1,4 IP2 (square). Measurements were performed at 20°C in buffer A containing 1 mM EDTA. (G) Ca2+ sensitivity of IRIS-1. Emission of IRIS-1 was measured in buffer A containing 1 mM HEDTA (circle) or 1 μM free Ca2+ (square). The free Ca2+ concentration was adjusted as described elsewhere (Michikawa et al., 1999). (H) pH sensitivity of IRIS-1. Three buffers with different pH (circle, pH 7.0; square, pH 7.4; triangle, pH 7.8) were prepared based on buffer A containing 1 mM EDTA. Error bars correspond to the SD (n = 3).