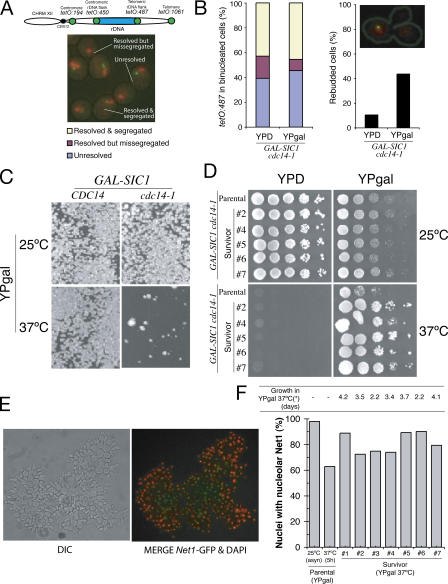
Figure 1.
Sic1-overexpression in cdc14-1 mutants creates a genetic “bottleneck” that corrects rDNA segregation defects. (A) Diagrammatic representation of the location of the chromosome tags along the right arm of chromosome XII used in this study. Representative micrograph of the different categories of tags scored in binucleated cells blocked by Cdc14 inactivation in B. (B) GAL-SIC1 cdc14-1 cells carrying chromosome tags in the centromere-distal flank of rDNA (tetO:487) were arrested in metaphase at 25°C and released to glucose (YPD; SIC1 overexpression off) or galactose (YPgal; SIC1 overexpression on) media at 37°C. The graphs show percentages of cells with segregated tags (left) and rebudding (right) 2 h after the metaphase release. In the top right corner, there is a picture of a cell that has entered a second cell cycle because of SIC1 overexpression but has failed to resolve the distal flank of the rDNA. (C) Yeast strains with the indicated genotypes were sonicated and plated onto YPgal medium (∼20 cells/cm2) and grown at the indicated temperatures for 4 d. A reduced growth of ∼1% of the total amount plated (survivors) is observed for GAL-SIC1 cdc14-1 at 37°C. (D) Different survivor colonies from C were grown in YPD at 25°C for ∼40 generations before they were 10-fold serially diluted, spotted onto different medium, and grown at the indicated temperatures for 3–4 d. (E) Representative micrograph of GAL-SIC1 cdc14-1 survivor cells expressing the nucleolar marker NET1-GFP (green) grown at 37°C in YPgal broth. DAPI is shown in red. Note that survivor cells grow as chains because of defects in cytokinesis. (F) Percentage of nuclei with Net1p-GFP signals in parental (GAL-SIC1 cdc14-1) and survivor (as in E) strains. Note that survivors show better segregation of Net1p-GFP.