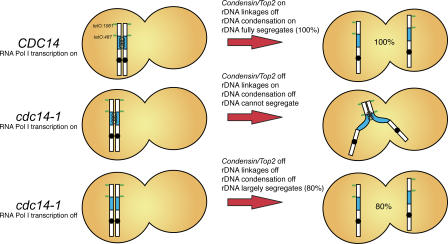
Figure 6.
RNA polymerase I transcription of rRNA genes and lack of rDNA condensation constrain chromosome XII disjunction in the absence of Cdc14. Diagrammatic representation of yeast chromosome XII with the rDNA array in blue and the position of various chromosome tags used in this study in green. Cdc14 activity during anaphase recruits condensin (D'Amours et al., 2004; Wang et al., 2004) to rDNA. RNA polymerase I transcription of chromosomal rRNA genes creates linkages between sister chromatids. Condensation of rDNA and removal of transcription-induced linkages are mediated by Cdc14, thus ensuring full segregation of chromosome XII (100% of cells). In the absence of Cdc14, neither rDNA condensation nor transcription-induced linkages are removed (middle); consequently, the mitotic disjunction of the distal regions of chromosome XII is prevented. When RNA polymerase I transcription is inactivated (and growth is supported by 35S RNA polymerase II–mediated transcription from a plasmid copy), transcription-induced linkages between sister chromatids do not arise and, consequently, distal regions of chromosome XII exhibit improved segregation (80% of cells are able to separate tags), even in the absence of Cdc14. Note that full segregation is not achieved (as in top row) because rDNA condensation is not induced. Our findings show that Cdc14-dependent rDNA condensation and resolution mechanisms are required to segregate the long arm of chromosome XII because of its size and the presence of linkages generated by the high transcription rates in rRNA genes.