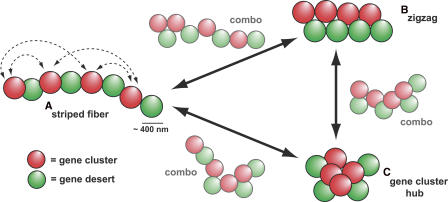
Figure 9.
Sequence-based, dynamic model of chromosome region structure. Dynamic conformations in interphase nuclei structurally manage the complex genetic information within a gene-poor chromosome region. Arrayed gene clusters (red) and deserts (green) of 200 kb–1 Mb act as building blocks for the formation of chromatin structures beyond the 30-nm fiber. Transient, probabilistic associations across a chromosome region, which are shown for gene clusters (dashed arrows), add up to predominant region conformations, such as a relatively linear striped fiber (A), a zigzag arrangement of gene clusters and deserts (B), and a centralized “gene cluster hub” with closely aggregated gene clusters (C). Intermediate combinations of the patterns (combo) further support dynamic transitions between one state and another (solid arrows). Though not shown, desert–desert and cluster–desert interactions also occur with defined probabilities.