Abstract
We report the supramolecular organization of killer Ig–like receptor (KIR) phosphorylation using a technique applicable to imaging phosphorylation of any green fluorescent protein–tagged receptor at an intercellular contact or immune synapse. Specifically, we use fluorescence lifetime imaging (FLIM) to report Förster resonance energy transfer (FRET) between GFP-tagged KIR2DL1 and a Cy3-tagged generic anti-phosphotyrosine monoclonal antibody. Visualization of KIR phosphorylation in natural killer (NK) cells contacting target cells expressing cognate major histocompatibility complex class I proteins revealed that inhibitory signaling is spatially restricted to the immune synapse. This explains how NK cells respond appropriately when simultaneously surveying susceptible and resistant target cells. More surprising, phosphorylated KIR was confined to microclusters within the aggregate of KIR, contrary to an expected homogeneous distribution of KIR signaling across the immune synapse. Also, yellow fluorescent protein–tagged Lck, a kinase important for KIR phosphorylation, accumulated in a multifocal distribution at inhibitory synapses. Spatial confinement of receptor phosphorylation within the immune synapse may be critical to how activating and inhibitory signals are integrated in NK cells.
Introduction
Natural killer (NK) cell activation is regulated by a balance between activating and inhibitory cell surface receptors (Vivier et al., 2004; Lanier, 2005b). Consistent with the missing self hypothesis (Ljunggren and Karre, 1990; Karre et al., 2005; Lanier, 2005a), NK cell cytotoxicity can be inhibited by engagement of inhibitory receptors specific for major histocompatibility complex (MHC) class I proteins, including killer Ig–like receptors (KIRs; Karlhofer et al., 1992; Yokoyama and Seaman, 1993; Colonna and Samaridis, 1995; Wagtmann et al., 1995). Initiation of the inhibitory signal upon ligand binding requires the phosphorylation of two tyrosine residues within immunoreceptor tyrosine-based inhibition motifs (ITIMs) in the cytoplasmic domain. These phosphorylated tyrosines act as a recruitment site for SH2 domain–containing tyrosine phosphatases, including Src homology protein tyrosine phosphatase (SHP) 1 or 2 (Burshtyn et al., 1996; Fry et al., 1996; Olcese et al., 1996; Burshtyn et al., 1999). Several signaling molecules involved in NK cell activation can be targets for SHP-1– and SHP-2–mediated dephosphorylation, including Zap70, Syk, PLCγ, LAT, and SLP76 (for review see Veillette et al., 2002). However, using a transfectant of YTS expressing KIR2DL1 fused to a substrate-trapping mutant of SHP-1, a guanine nucleotide exchange factor that regulates the actin cytoskeleton, Vav-1, was the only protein detected as a direct substrate for SHP-1 (Stebbins et al., 2003). Downstream, inhibitory KIR2DL1 signaling prevents the assembly of a large complex of cytoskeletal-linked proteins required for cytotoxicity (Krzewski et al., 2006).
KIR phosphorylation after engagement of MHC class I protein on target cells has proved difficult to detect biochemically and has in some cases required addition of a phosphatase inhibitor, pervanadate, to facilitate its detection (Faure et al., 2003). The most likely explanation for this is that only a small fraction of KIR is phosphorylated at any given moment. Thus, determining where and when inhibitory KIR signaling occurs is an essential next stage toward understanding how the balance of activating and inhibitory signals is assessed during NK cell surveillance.
KIR and their corresponding MHC class I ligands, as well as many other receptor/ligand pairs, have been shown to cluster at the immunological synapse (IS) between NK cells and other cells (Davis et al., 1999; Carlin et al., 2001; Vyas et al., 2001, 2002; Orange et al., 2003; Borg et al., 2004). However, whether there is an importance in the segregation and patterning of proteins at an inhibitory NK cell IS, e.g., in influencing downstream signaling, remains unclear (Davis and Dustin, 2004). We set out to determine the supramolecular organization of the first step in inhibitory receptor signaling, phosphorylation of the cytoplasmic ITIMs of KIR2DL1. Förster resonance energy transfer (FRET) involves the nonradiative transfer of energy from an excited donor fluorophore to a nearby acceptor and can be used to detect macromolecular associations within cells on the nanometer scale (Wu and Brand, 1994). Here, we image KIR phosphorylation at the NK cell IS using fluorescence lifetime imaging (FLIM) to report FRET. Rather than a small fraction of KIR being phosphorylated homogeneously across the IS, we unexpectedly observed that KIR phosphorylation is spatially restricted to discrete domains or microclusters within the IS.
Results
Imaging KIR phosphorylation at the inhibitory NK cell IS
To visualize KIR signaling, we used a generic anti-phosphotyrosine mAb labeled with Cy3 as the acceptor for FRET from the donor GFP tagged to the cytoplasmic portion of the NK inhibitory receptor KIR2DL1. FRET will only be detected if the spatial separation of GFP and Cy3 fluorophores is no more than 9 nm (Ng et al., 1999); thus, FRET will occur only when the anti-phosphotyrosine mAb is extremely close, i.e., bound, to KIR2DL1-GFP. The most robust way to detect FRET is through detecting a decrease in the fluorescence lifetime, τ, of the donor fluorophore, in this case, GFP (Bastiaens and Squire, 1999). Hence, KIR phosphorylation at the NK cell IS can be detected by comparing the fluorescence lifetime of GFP-tagged KIR2DL1 in unstained cell conjugates (donor only [D]) with the fluorescence lifetime of GFP-tagged KIR2DL1 in conjugates stained with Cy3-labeled anti-phosphotyrosine (donor in the presence of acceptor [DA]; Fig. 1).
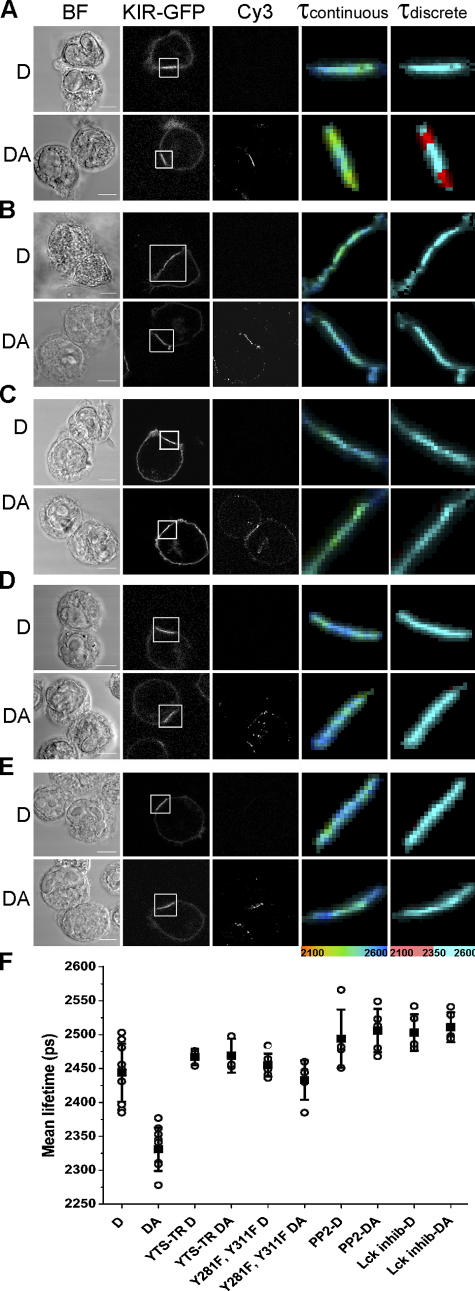
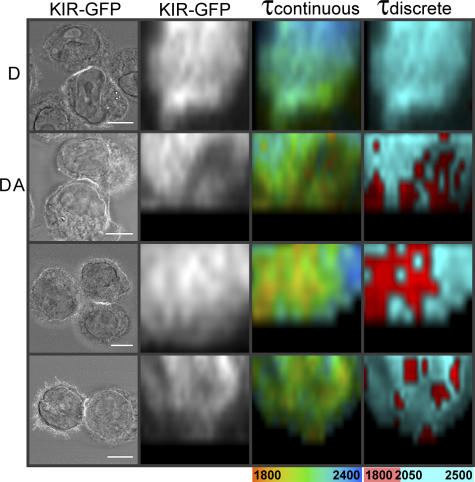
Figure 1.
Imaging KIR2DL1 phosphorylation at the inhibitory NK cell IS. YTS cells expressing full-length KIR2DL1-GFP (A), a truncated ITIM-less KIR2DL1-GFP (YTS-TR; B), or KIR2DL1-GFP (Y281F and Y311F; C) in which the two ITIM tyrosines had been changed to phenylalanine were coincubated with 221/Cw6 target cells and then fixed and stained. YTS/KIR2DL1-GFP cells were treated with 5 μM PP2 (D) or 5 μM Lck-specific inhibitor (E) before coincubation with target cells. KIR phosphorylation is detected by a reduction in the fluorescence lifetime (τ) in picoseconds of KIR2DL1-GFP in the presence of the Cy3-tagged anti-phosphotyrosine mAb acceptor (DA) compared with an unstained sample (D). Images show the fluorescence lifetimes (within the area of the IS; white box) mapped to a continuous scale (τcontinuous; 2,100–2,600 ps) or a discrete scale (τdiscrete), where the breakpoint at 2,350 ps corresponds to 5% EFRET. Images shown are representative of at least two experiments and a total of 8–15 cell conjugates imaged in each condition. Bars, 8 μm. (F) Mean lifetime ± SD of KIR2DL1-GFP was calculated by selecting the region of the synapse for unstained cells (D) and cells stained with Cy3-tagged anti-phosphotyrosine mAb (DA) for each condition. Values shown in the graph are the mean lifetimes across the synapse, which would therefore include a mixture of pixels where FRET was present and absent.
An accumulation of KIR2DL1-GFP and phosphotyrosine is clearly visible at the IS between YTS/KIR2DL1-GFP and 221/Cw6. The fluorescence lifetime of GFP is reduced in samples stained with anti-phosphotyrosine compared with unstained samples (Fig. 1 A). The mean fluorescence lifetime where KIR clustered at the IS in conjugates stained for phosphotyrosine (DA) was 5–10% lower than unstained control cells (D; Fig. 1 F). The fluorescence lifetime of GFP was not reduced in anti-phosphotyrosine–stained samples of YTS/KIR2DL1-GFP and 221/Cw3 that express a class I MHC protein not recognized by KIR2DL1 (unpublished data).
However, KIR2DL1 does not cluster at these synapses with target cells lacking a cognate MHC ligand. Thus, to further test whether the observed decrease in GFP fluorescence lifetime at the IS between YTS/KIR2DL1-GFP and 221/Cw6 specifically reported KIR phosphorylation, a transfectant expressing a truncated ITIM-less KIR2DL1-GFP (YTS-TR) was used, where GFP was placed just upstream of the membrane-proximal ITIM and the rest of the KIR cytoplasmic tail deleted. Truncated KIR2DL1-GFP still clustered at the IS with target cells expressing cognate MHC protein (Fig. 1 B). Thus, signaling through ITIMs, or any other possible signaling motifs, in the cytoplasmic tail of KIR is not absolutely required to cluster KIR at the IS, consistent with previous observations (Fassett et al., 2001; Standeven et al., 2004).
There was clearly anti-phosphotyrosine staining at the synapse involving YTS expressing truncated KIR2DL1, as expected because KIR is not the only protein to be phosphorylated at a synapse, and indeed the lack of KIR signaling would allow a cytolytic synapse to persist. However, FRET could not be detected at the IS despite truncated KIR2DL1-GFP being clustered at the IS, where phosphotyrosine is also present (Fig. 1, B and F). To control for the possibility that our methodology would be sensitive to phosphorylated proteins that might associate with the cytoplasmic tail independently of ITIM phosphorylation, we also used a transfectant expressing KIR2DL1-GFP (Y281F, Y311F), in which the two ITIM tyrosines had been specifically altered to phenylalanine. Again, FRET could not be detected in synapses involving transfectants expressing this point-mutated ITIM-less KIR2DL1-GFP (Fig. 1, C and F), despite this receptor being clustered at the IS, where phosphotyrosine is also present.
Together, these controls confirm that our methodology accurately reveals the phosphorylation of KIR2DL1 at the IS and does not report on other phosphorylated proteins that might happen to be in close proximity to KIR2DL1 nor on any proteins that might be bound to the cytoplasmic tail of KIR2DL1 irrespective of ITIM phosphorylation. It is possible, however, that phosphotyrosine-containing proteins specifically recruited to phosphorylated ITIMs also contribute to the FRET signal seen, but this will still indicate active KIR2DL1 signaling, as is our intent. Thus, using FLIM to measure FRET between a GFP-tagged receptor and a Cy3-labeled generic anti-phosphotyrosine mAb is a sensitive technique for detecting specific receptor signals at an IS. This generic method could be applied to image phosphorylation of any receptor at the IS and is especially useful for probing phosphorylated sites for which specific mAb are not available.
KIR phosphorylation requires an Src family kinase
We next imaged whether or not KIR phosphorylation would persist after treatment with an Src family kinase inhibitor, PP2, or an Lck-specific inhibitor (Burchat et al., 2000). KIR2DL1-GFP clustered at the IS after treatment of NK cells with either inhibitor (Fig. 1, D and E), indicating that Src family kinase–mediated signaling is not necessary for KIR clustering, consistent with previous studies (Standeven et al., 2004). Anti-phosphotyrosine staining was also still apparent at the IS, consistent with phosphorylation of some proteins at the IS being mediated by other kinases. Despite KIR2DL1 receptor clustering and anti-phosphotyrosine staining, FRET was not detected in the presence of either PP2 or the Lck-specific inhibitor (Fig. 1, D–F). Therefore, the activity of Src family protein tyrosine kinases, and specifically Lck, are necessary for KIR phosphorylation at the IS.
KIR phosphorylation is spatially restricted to the IS and lasts several minutes
To examine the dynamics of KIR phosphorylation at the inhibitory NK cell IS, we imaged conjugates fixed after different times of coincubation. The fluorescence lifetime of KIR2DL1-GFP decreased in the presence of Cy3-tagged anti-phosphotyrosine mAb across all the times examined (Fig. 2). KIR signaling is thus sustained for some minutes, perhaps ensuring ongoing interruption of competing activating signals. Changes in the FRET efficiency relate to the relative extent of KIR phosphorylation within the synapse (Fig. 2, A–C; right). The extent of FRET detected at individual synapses varied, such that a relatively large degree of KIR phosphorylation occurred at some synapses, whereas little occurred at others. The mean FRET efficiency across several synapses was 4.3, 5.2, and 2.8% after 5, 10, and 20 min of coincubation, respectively (Fig. 2 D). Although the FRET efficiency does not directly report the number of activated KIRs, these numbers do suggest that only a surprisingly small fraction of KIR is phosphorylated at a given moment, explaining why KIR phosphorylation has been difficult to detect biochemically.
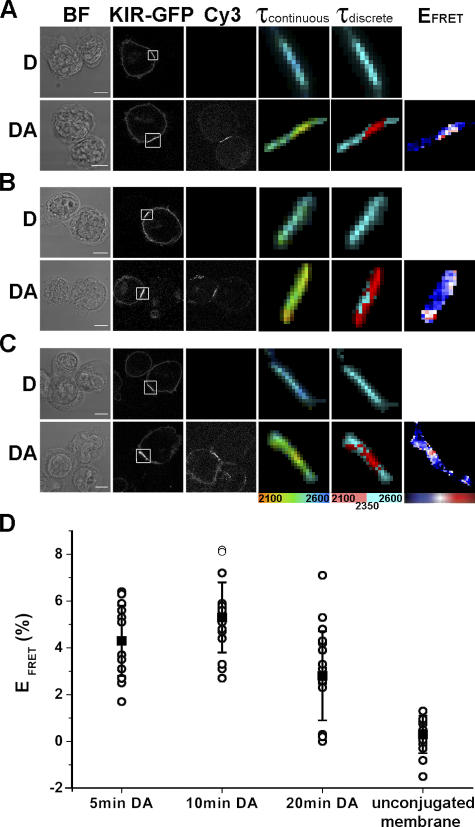
Figure 2.
KIR2DL1 phosphorylation is sustained at the inhibitory NK cell IS. YTS/KIR2DL1-GFP cells were coincubated with 221/Cw6 for 5 min (A), 10 min (B), or 20 min (C) and then fixed and stained with Cy3-labeled anti-phosphotyrosine mAb. Images show the fluorescence lifetime (within the area of the IS; white box) mapped to a continuous scale (τcontinuous; 2,100–2,600 ps) or a discrete scale (τdiscrete), where the breakpoint at 2,350 ps corresponds to 5% EFRET. FRET efficiencies were calculated for each pixel as EFRET (%) = 1 − τDA/τD × 100, where τDA is the fluorescence lifetime of the donor in the presence of the acceptor and τD is the mean fluorescence lifetime of the donor in the absence of the acceptor (unstained controls). Color scale shown covers EFRET 0–15% for 5 and 10 min and 0–10% for 20 min. Images shown are representative of at least three experiments and a total of 15–20 cell conjugates imaged in each condition. Bars, 8 μm. (D) Mean EFRET ± SD at the IS or in the unconjugated membrane for each cell conjugate were calculated, where τDA is the mean fluorescence lifetime of the donor at the IS in the presence of the acceptor and τD is the mean fluorescence lifetime of the donor at the IS in the absence of the acceptor (unstained controls).
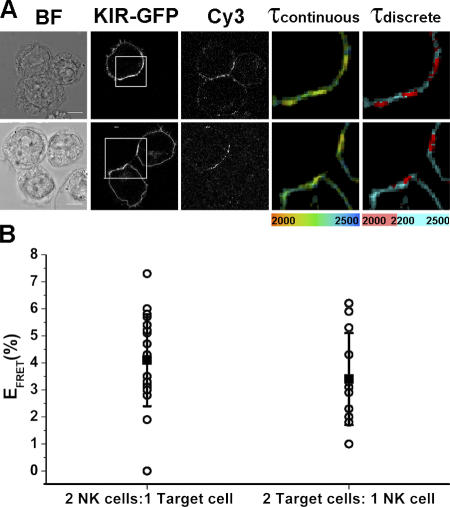
Interestingly, there was no decrease in the fluorescence lifetime outside where KIR2DL1-GFP clustered at the intercellular contact (Fig. 2 D). Thus, KIR phosphorylation occurred only locally at the IS, in contrast to EGF receptor signaling, for example, which rapidly spreads throughout the cell membrane (Verveer et al., 2000). Even when a single NK cell engages two resistant target cells, KIR phosphorylation is restricted to the intercellular contact with both targets (Fig. 3 A, top) and does not spread to the remaining, unconjugated membrane. By not spreading KIR phosphorylation outside the IS, NK cell inhibition can be restricted to one target cell while maintaining an effective surveillance of another conjugated cell, allowing NK cells to survey both susceptible and resistant target cells simultaneously and respond appropriately (Eriksson et al., 1999). We also found that a single target cell can effectively trigger KIR phosphorylation when bound to two NK cells simultaneously (Fig. 3 A, bottom), ensuring that accumulation of human leukocyte antigen (HLA)–C at one synapse would not make the target cell susceptible to attack by another bound NK cell.
Figure 3.
KIR phosphorylation occurs specifically at the IS. YTS/KIR2DL1-GFP cells were coincubated with 221/Cw6. (A) Multiple cell conjugates consisting of either a single NK cell in conjugation with two target cells (top) or a single target cell in conjugation with two NK cells (bottom) were imaged by FLIM. The fluorescence lifetime, within the area of the IS (white box), of KIR2DL1-GFP in the presence of the acceptor is shown on a continuous scale (τcontinuous; 2,000–2,500 ps) or a discrete scale (τdiscrete), where the breakpoint at 2,200 ps corresponds to 5% EFRET. Bars, 8 μm. (B) The range in EFRET at synapses formed by cells involved in multiple contacts is shown and is similar to the range of EFRET observed in cells making single contacts.
KIR phosphorylation occurs within microclusters
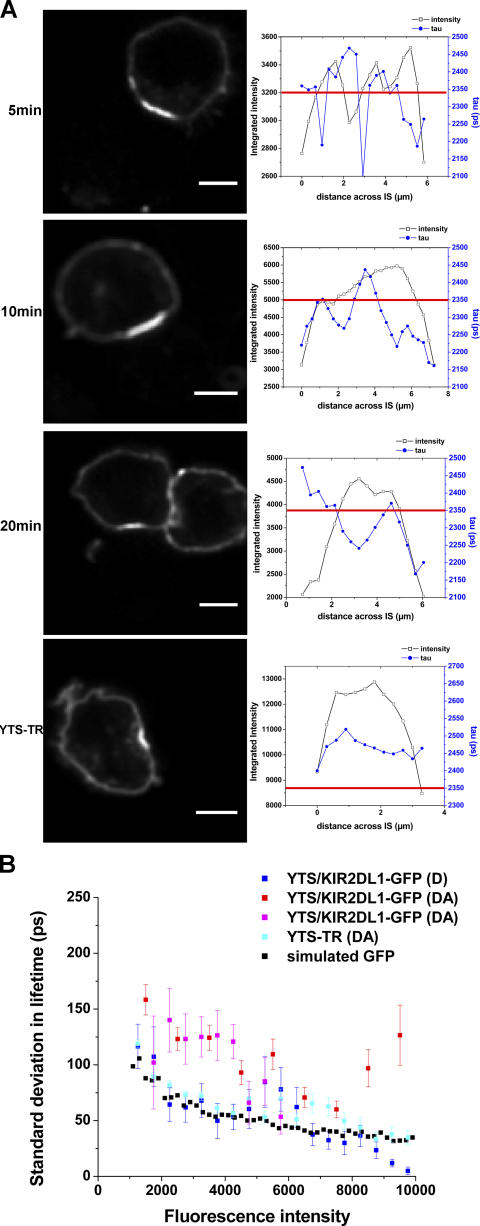
Analysis of the intensity and fluorescence lifetime values across the synapse revealed discrete domains with decreased lifetime within the aggregate of KIR2DL1-GFP at the IS (Fig. 4 A). Such “microclusters” of phosphorylated KIR did not specifically colocalize with an increased concentration of KIR2DL1-GFP, as determined by comparing the location of FRET (decreased lifetime) with the intensity of GFP (Fig. 4 A, top three panels). Domains of decreased fluorescence lifetime were not observed at synapses with cells expressing KIR-GFP lacking ITIMs (KIR-TR; Fig. 4 A, bottom). To statistically assess whether the observed microclusters with decreased lifetime could arise from natural statistical variation in the measured fluorescence lifetime, we quantitatively analyzed the standard deviation in the lifetime of KIR2DL1-GFP as a function of intensity compared with a simulated dataset, as previously described (Treanor et al., 2005). Effectively, the simulated data establishes a baseline in the variability of measured lifetime values that arise as a consequence of noise in the photon-counting process and not due to any real variation in the fluorescence lifetime of the sample. The standard deviation of the fluorescence lifetime of KIR2DL1-GFP at the inhibitory synapse in the presence of an acceptor is significantly greater than the simulated GFP or that observed for the ITIM-less KIR2DL1-GFP (Fig. 4 B), confirming that there are true differences in the fluorescence lifetime of KIR2DL1-GFP. Our analysis does not allow us to precisely determine the size of microclusters of phosphorylated KIR. However, taking these data together, structure in the organization of phosphorylated KIR must occur on a scale close to the limit of the spatial resolution of our method, i.e., 1 μm. This scale could represent the size of microclusters themselves or a distance between smaller microclusters. Very small microclusters, e.g., below the limit of resolution for optical microscopy (∼0.4 mm), would be less visible. Large clusters of phosphorylated KIR, i.e., 3–4 μm, were sometimes seen, which may result from an aggregation of smaller microclusters. For comparison, the size of the cluster of KIR2DL1-GFP was 5–8 μm.
Figure 4.
KIR phosphorylation occurs in discrete microclusters. (A) KIR2DL1-GFP fluorescence intensity and fluorescence lifetime were analyzed across the region of clustered KIR (left) by plotting (right) the intensity (black line, left scale) and fluorescence lifetime (blue line, right scale) for each pixel across the synapse. For reference, the lifetime representing where 5% FRET efficiency occurred for each sample is also depicted (red line). Data shown are for YTS/KIR2DL1-GFP cells coincubated with 221/Cw6 for 5, 10, or 20 min, or YTS-TR cells. Data are representative of at least three experiments and a total of 15–20 cell conjugates imaged in each condition. (B) SD in fluorescence lifetime as a function of intensity for a simulated GFP dataset and representative cell conjugates of YTS/KIR2DL1-GFP and 221/Cw6 (D and two examples of DA) or YTS-TR and 221/Cw6 (DA). Bars, 8 μm.
3D imaging of FRET was obtained by fluorescence lifetime images being acquired every 0.5 μm throughout the conjugate, and the en face fluorescence intensity and lifetime at the IS was reconstructed (Fig. 5). Discrete regions of decreased fluorescence lifetime within the cluster of KIR were clearly visible. Thus, KIR signaling does not occur uniformly across the IS and, although there is potentially significant KIR signaling occurring in other areas of the synapse, the majority of KIR signaling appears spatially confined to discrete microclusters within the larger aggregation of receptor at the IS.
Figure 5.
3D FLIM of FRET reveals microclusters of signaling at the inhibitory NK cell IS. 3D imaging of FRET was obtained by FLIM images being acquired every 0.5 μm throughout the conjugate. En face reconstructions are shown of KIR2DL1-GFP intensity and τ (in ps) plotted on a continuous scale (1,800–2,400 ps) and a discrete scale where the breakpoint of 2,050 ps represents the point corresponding to 5% EFRET. Images are representative of at least three experiments and a total of 30 cell conjugates. Bars, 8 μm.
Microclusters of NK cell Lck accumulate at inhibitory synapses
We next set out to test for the presence of microclusters of signaling in live cell–cell conjugates. Visualization of KIR phosphorylation at the NK cell IS required accurate detection of low levels of FRET. Single photon counting is the method of choice for the most accurate measurements of fluorescence lifetime. However, single photon–counting FLIM is inherently slow (Suhling et al., 2005) and thus it is not possible to extend our methodology to detect KIR phosphorylation in fast-moving live cell interactions. Instead, because we found that, consistent with previous studies (Binstadt et al., 1996), Lck was necessary for KIR phosphorylation (Fig. 1), we set out to determine whether the kinase itself would accumulate in microclusters at the inhibitory NK cell IS.
To assess the cellular distribution of Lck in live cells, YTS/KIR2DL1 cells were transfected to express Lck conjugated to monomeric YFP (mYFP). Western blotting confirmed the presence of Lck-mYFP at the expected size in these transfectants (unpublished data). Confirming that the chimera Lck-mYFP could be functional, expression of Lck-mYFP in JCam1.6 restored the ability of this Lck-deficient cell line to flux calcium after anti-CD3 mAb stimulation (unpublished data), consistent with previous observations (Ehrlich et al., 2002). Expression of Lck-mYFP did not alter the cytolytic response of YTS/KIR2DL1, as target cells expressing a noncognate MHC class I protein (221/Cw3) were still lysed by this transfectant, whereas target cells expressing a cognate MHC class I protein (221/Cw6) were resistant (unpublished data).
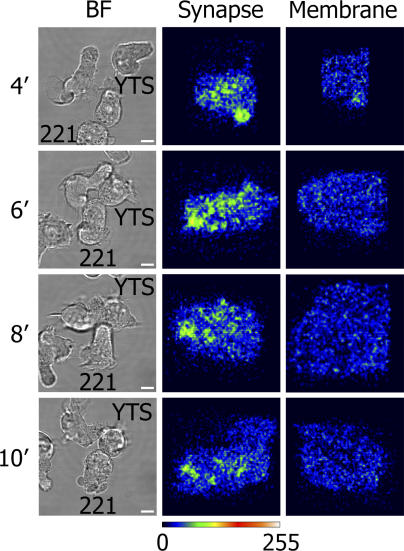
In images, Lck-mYFP was localized at the YTS/KIR2DL1 plasma membrane as well as in an intracellular pool, as previously observed in peripheral blood NK cells (Vyas et al., 2002) and T cells (Ehrlich et al., 2002). Soon after contact with target cells, small clusters of membrane-associated Lck are clearly visible at the IS (Fig. 6), consistent with previous studies observing clusters of Lck at NK cell synapses in fixed cells (Vyas et al., 2001). Small clusters of Lck-YFP were sustained for several minutes at the inhibitory NK cell IS (Fig. 6). The presence of small clusters of Lck being sustained at the inhibitory NK cell IS coupled with observations of microclusters of phosphorylated KIR is consistent with Lck being necessary for KIR phosphorylation and demonstrates further that NK cell signaling occurs within microclusters.
Figure 6.
Lck is distributed in microclusters at inhibitory NK cell synapses. Live cell imaging of YTS/KIR2DL1 transfected to express Lck-mYFP in the presence of 221/Cw6 target cells was performed by resonance scanning confocal microscopy. Brightfield images of a representative conjugate are shown (BF), with the corresponding reconstruction of the IS viewed en face and colored according to intensity (synapse) and, for comparison, unconjugated cell membrane away from the IS (membrane). Images are shown at 4, 6, 8, and 10 min after initial contact between cells. Data are representative of six time-lapse 3D series obtained from three independent experiments. Bars, 10 μm.
Discussion
We visualized signaling at an IS using FLIM to report FRET between GFP-tagged KIR and a fluorophore-tagged general anti-phosphotyrosine mAb. This technology can be applied to image the phosphorylation of any specific receptor at an intercellular contact. This methodology is of particular use where phosphospecific mAbs are not available and could be extended toward developing rapid screens for protein phosphorylation. Using this methodology, we found that KIR signaling occurs within discrete microclusters within the larger aggregation of protein at the NK cell IS. Imaging of T cell signaling events recently revealed microclusters of TCR colocalized with activated forms of Lck, ZAP-70, and LAT at the contact between a T cell and a lipid bilayer (Campi et al., 2005; Trautmann, 2005; Yokosuka et al., 2005; Saito and Yokosuka, 2006), which builds on earlier work showing TCR signaling clusters assembled at contacts with antibody-coated coverslips (Bunnell et al., 2002). Thus, our observations extend the generality of microcluster-mediated signaling by demonstrating their relevance to human NK cell inhibitory signaling.
What then, is restricting KIR phosphorylation to discrete microclusters? Although the Singer-Nicolson fluid mosaic model has formed the basis of our understanding of the cell membrane for the past three decades, relatively recent evidence suggests the organization of both proteins and lipids to be much more complex than this (Engelman, 2005). Here, it is unlikely that lipid rafts facilitate the formation of microclusters of inhibitory signaling, as KIR2DL1 is excluded from such domains and in fact signals to block an accumulation of lipid rafts to the IS (Lou et al., 2000; Fassett et al., 2001). Alternatively, specific protein–protein interactions could create microclusters of KIR2DL1 phosphorylation or they might be a result of “membrane-skeleton corralling,” i.e., by the cytoplasmic domains of transmembrane proteins colliding with the cytoskeleton (Kusumi et al., 2005). Such temporary confinement would be further enhanced by oligomerization, and indeed metal ions, known to dimerize KIR2DL1 in solution (Fan et al., 2000), have been found to be necessary for KIR function and phosphorylation (Rajagopalan et al., 1995; Faure et al., 2003).
It is perhaps surprising that spatially confined inhibitory KIR signals are effective in inhibiting NK cell responses. However, the observation that KIR need not be continuously actively inhibiting over the entire synapse is consistent with the observation that a chimera of KIR tagged with an extracellular GFP was able to efficiently inhibit NK cell responses without being significantly clustered at the IS (Borszcz et al., 2003). In general, spatially confined recruitment of kinases to the NK cell synapse may facilitate the transphosphorylation of numerous NK cell receptors within a microcluster. For example, trapped Lck, as recently visualized at the interface between a T cell and a mAb-coated coverslip (Douglass and Vale, 2005), could facilitate transphosphorylation between KIR and other NK cell receptors within discrete microclusters.
Thus, one specific hypothesis for an efficient mechanism of KIR-mediated inhibition would be that activating NK cell receptors recruit Lck, which is then able to transphosphorylate local KIR if they are engaged by HLA-C on the opposing cell. This model therefore requires that signaling by NK cell–activating receptors occurs within microclusters, analogous to TCR signals. This could be caused by NK cell–activating ligands being organized into specific microdomains on the surface of the opposing target cell (Eleme et al., 2004). Thus, in this model, the spatial confinement of inhibitory receptor phosphorylation serves to focus inhibitory action, e.g., the dephosphorylation of Vav-1 (Stebbins et al., 2003), to specific sites where the competing activating signals are triggered.
Broadly, it would previously have been expected that KIR signaling occurs homogeneously across the cluster of KIR at the IS. Thus, the data presented here seeds new research into how the spatial confinement of receptor phosphorylation could influence the integration of activating and inhibitory signals by NK cells.
Materials and methods
Cells
A transfectant of the MHC-deficient human B-lymphoblastoid cell line, 721.221 expressing HLA-Cw6 (221/Cw6) has been described (Davis et al., 1999). YTS, a subclone of the human tumor line YT, expressing KIR2DL1 (YTS/KIR2DL1; Cohen et al., 1999), COOH-terminal GFP-tagged KIR2DL1 (YTS/KIR2DL1-GFP; Borszcz et al., 2003), or a truncated ITIM-less KIR2DL1-GFP (YTS-TR; Standeven et al., 2004) where GFP was placed just upstream of the membrane-proximal ITIM and the rest of the KIR was cytoplasmic tail deleted have also been described. A mutant of KIR2DL1-GFP (Y281F and Y311F) in which the two ITIM tyrosines were mutated to phenylalanine was generated in pBABE (QuikChange Mutagenesis; Stratagene). The membrane-distal tyrosine was mutated to phenylalanine first using the forward primer 5′-GATATCATCGTGTTCACGGAACTTCC-3′ and its reverse complement. The membrane-proximal tyrosine was then mutated to phenylalanine using the forward primer 5′-CCTCAGGAGGTGACATTCACACAGTTGAATC-3′ and its reverse complement. The fidelity of the construct was confirmed by sequencing and expressed in YTS cells by retroviral transduction as described previously (Borszcz et al., 2003).
For Lck-mYFP, human Lck was amplified by PCR from cDNA synthesized from PBL using primers 5′-CCCAAGCTTGCC ACCATGGGCTGTGGCTGCAGCTC-3′, including a HindIII restriction site, and 5′-GCGGTACCCCAGGCTGAGGCTGGTACTGGCCCTC-3′ to remove the stop codon and include a KpnI restriction site. The PCR product was first cloned into pCR2.1-TOPO (Invitrogen), and the correct sequence was confirmed and subcloned into the mammalian expression vector pcDNA 3.1/Hygro containing mYFP, i.e., with substitutions S65G, S72A, T203Y, and A206K (a gift from R. Tsien, University of California, San Diego, La Jolla, CA). This resulted in a construct encoding Lck and mYFP connected by an 11-amino-acid linker (Gly-Val-Pro-Ser-Ser-Asp-Pro-Pro-Val-Ala-Thr). YTS-KIR2DL1 and a variant of Jurkat lacking Lck, JCam1.6 (American Type Culture Collection), were each transfected to express Lck-mYFP by electroporation. Transfectants were grown in 0.8 mg/ml hygromycin, and Lck-mYFP–expressing cells were sorted by flow cytometry (FACSDiva; Becton Dickinson).
Cell conjugation and staining
For cell conjugation, 5 × 105 YTS/KIR2DL1, YTS/KIR2DL1-GFP, YTS-TR, or YTS-KIR2DL1 (Y281F and Y311F) was mixed with 5 × 105 221/Cw6 cells in 50 μl warm media and incubated at 37°C/5% CO2 for the indicated time. Cell conjugates were then fixed in 100 μl buffer containing paraformaldehyde and saponin (Cytofix/Cytoperm; Becton Dickinson) for 15 min at 4°C followed by 5 min at room temperature. Cells were washed in 0.1% Tween-20/PBS and blocked in 100 μl buffer containing saponin (Perm/Wash; Becton Dickinson) with 5% horse serum/3% BSA for 30 min at 4°C. Cell conjugates were stained in 100 μl of 10 μg/ml anti-phosphotyrosine mAb (clone 4G10; Upstate Biotechnology) tagged with Cy3 (Cy3/mAb ratio 8:1) for 2 h at 4°C. After washing, cell conjugates were gently resuspended and ∼8 μl was placed between a microscope slide (thickness 1 mm; Becton Dickinson) and a glass coverslip (thickness No. 1.5; Becton Dickinson).
Inhibitors
The Src family tyrosine kinase inhibitor PP2 (Calbiochem) and the Lck-specific inhibitor, 7-Cyclopentyl-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-ylamine (Burchat et al., 2000; Sigma-Aldrich) were diluted to 5 μM in cell media. YTS transfectants were preincubated with inhibitors for 1 h at 37°C/5% CO2 before mixing with target cells for 10 min in the continuing presence of the inhibitor.
FLIM to measure FRET
FLIM was performed using an inverted scanning confocal microscope (DMIRE2/TCS SP2; Leica Microsystems Ltd) with a 63× oil immersion (NA 1.3). Single-photon excitation was achieved using a solid-state, diode-pumped, frequency-doubled Nd:YAG laser (Millennia; Spectra-Physics Ltd) to pump a mode-locked frequency-doubled Ti:Sapphire laser (Tsunami; Spectra-Physics Ltd) that provided optical pulses of 100-fs full width at half maximum at a repetition rate of 80 MHz. The laser was double passed through a glass block (total interaction length 130 cm) to stretch the pulse to a final width of 13 ps. The optimal excitation wavelength to excite the donor with minimal direct excitation of the acceptor was determined to be 470 nm. Fluorescence emission of KIR2DL1-GFP was collected using a narrow bandpass filter (515 ± 15 nm) to limit detection to only the donor fluorophore (GFP) and prevent contamination from acceptor (Cy3) emission. Fluorescence intensity images for Cy3-tagged anti-phosphotyrosine mAb were collected from 600–700 nm to limit bleed-through of the donor GFP fluorescence, resulting in images that are much dimmer than if the whole Cy3 emission spectrum was collected.
The fluorescence lifetime of GFP-tagged KIR2DL1 was measured using time-correlated single photon counting (SPC-730; Becker & Hickl GmbH). Laser power was adjusted to give a mean photon count rate of ∼1 × 105 counts/s, and fluorescence lifetime images were acquired over 300 s. Fluorescence lifetimes were calculated for all pixels in the field of view (128 × 128 pixels; SPCImage). As the fluorescence intensity in the unconjugated (nonsynapse) membrane was very low, it was necessary to bin all photons from this region to accurately calculate the fluorescence lifetime. To achieve this, an in house–written fluorescence-decay program (written in Labview [National Instruments]) was used. The fluorescence lifetime for the synapse was also calculated using this program and agreed well with the fluorescence lifetime calculated using SPCImage.
FRET efficiency images were calculated such that the FRET efficiency, EFRET = 1 − τDA/τD, where τDA is the pixel-by-pixel fluorescence lifetime of the donor in the presence of the acceptor and τD is the mean fluorescence lifetime of the donor at the IS in the absence of the acceptor for all cells imaged (unstained controls). Mean FRET efficiencies at the IS were calculated where τDA is the mean fluorescence lifetime of the donor in the presence of the acceptor and τD is the mean fluorescence lifetime of the donor in the absence of the acceptor.
3D imaging of FRET was obtained by fluorescence lifetime images being acquired every 0.5 μm throughout the conjugate. An in house–written MatLab program was used to generate the en face fluorescence lifetime data for those synapses that were orientated vertically or horizontally to the x axis of the image. To select the pixels in the synapse volume, a region of interest was first created around a synapse in one of the image slices from the stack. This produced a 4D matrix consisting of the three spatial dimensions of the synapse and the lifetime intensity information for each pixel. The lifetime intensity information was then integrated along one of the spatial dimensions, x or y, i.e., perpendicular to the synapse region selected, effectively collapsing the volume to produce a 2D lifetime image (x or y and z with lifetime data). This dataset was then used to produce the en face intensity and fluorescence lifetime images (SPCImage fitting software). Similar images, demonstrating microclusters of signaling, were also obtained using commercially available 3D rendering software (Volocity; Improvision) to create the en face image of optical sections individually processed in SPCImage (unpublished data).
Image presentation
As fluorescence lifetime images were limited to 128 × 128 pixels, the limit in resolution of the photon counting detector, it was necessary to apply an interpolation method to obtain enlarged images of synapses at suitable resolution and size for publication. For this, the “nearest neighbor” interpolation method (Photoshop 7; Adobe) was applied, which sets the value (or color, in this case) of an interpolated point to the value of the nearest existing data point, effectively making the pixels bigger. For example, to enlarge 200%, one pixel will be enlarged to a 2 × 2 area of four pixels with the same color as the original pixel. This is the most appropriate method for interpolation of indexed images, i.e., images that map pixel values to colors, as it does not change the color information of the image and does not introduce any anti-aliasing, which would make edges appear smoother by averaging out pixels. An exception is that for reconstruction of the en face synapse (shown in Fig. 5) the bicubic method of interpolation was applied. We carefully confirmed that the images of synapses did not appear different by applying this procedure, and analysis of fluorescence lifetime data was always performed on the raw, noninterpolated data.
Variability in FLIM
It is clear that random errors caused by instrumental drift or subtle variation in the biological sample will effect different lifetime images far more so than within a single image. It has been specifically calculated that interimage differences, using a frequency-based FLIM methodology, are typically one order of magnitude greater than intraimage variations (Hanley et al., 2001). We have addressed this in three ways: (1) In all experiments, we compared image data taken from one specific experiment performed over one single day. (2) We confirmed that the presence or absence of FRET was consistent in each sample over multiple independent experiments. (3) The “breakpoint” in all images using a discrete scale for the fluorescence lifetime is specifically set at the point where FRET efficiency is 5% for that experiment.
Live cell imaging
YTS/KIR2DL1 cells transfected to express Lck-mYFP and 221/Cw6 target were mixed in a glass-bottomed microscope chamber (Nunc) containing 200 μl of warm RPMI media. Live cell conjugates were imaged by resonance scanning confocal microscopy (DMIRE2/TCS SP2 RS) using a 63× water-immersion objective (NA 1.2). The microscope stage was housed within an environmental chamber (Solent Scientific) maintained at 37°C/5% CO2. mYFP was excited using the 514-nm line of an argon laser. 3D views of cell conjugates and en face views of the synapse were reconstructed using Volocity.
Acknowledgments
We thank Björn Önfelt for discussions.
We acknowledge financial support from a Department of Trade and Industry Beacon award (QCBB/C/007/00007), the Medical Research Council (grant GO500563), the Chemical Biology Centre, the Biotechnology and Biological Sciences Research Council (grant BB/C512896/1), the European Community (Framework VI Integrated Project “Integrated technologies for in vivo molecular imaging” contract LSHG-CT-2003-503259), the Canadian Institutes of Health Research (grant CIHR MOP 36344), and a Lister Institute Research Prize (to D.M. Davis).
Abbreviations used in this paper: FLIM, fluorescence lifetime imaging; FRET, Förster resonance energy transfer; HLA, human leukocyte antigen; IS, immunological synapse; ITIM, immunoreceptor tyrosine-based inhibition motif; KIR, killer Ig–like receptor; MHC, major histocompatibility complex; mYFP, monomeric YFP; NK, natural killer; SHP, Src homology protein tyrosine phosphatase.
References
- Bastiaens, P.I., and A. Squire. 1999. Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol. 9:48–52. [DOI] [PubMed] [Google Scholar]
- Binstadt, B.A., K.M. Brumbaugh, C.J. Dick, A.M. Scharenberg, B.L. Williams, M. Colonna, L.L. Lanier, J.P. Kinet, R.T. Abraham, and P.J. Leibson. 1996. Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity. 5:629–638. [DOI] [PubMed] [Google Scholar]
- Borg, C., A. Jalil, D. Laderach, K. Maruyama, H. Wakasugi, S. Charrier, B. Ryffel, A. Cambi, C. Figdor, W. Vainchenker, et al. 2004. NK cell activation by dendritic cells (DCs) requires the formation of a synapse leading to IL-12 polarization in DCs. Blood. 104:3267–3275. [DOI] [PubMed] [Google Scholar]
- Borszcz, P.D., M. Peterson, L. Standeven, S. Kirwan, M. Sandusky, A. Shaw, E.O. Long, and D.N. Burshtyn. 2003. KIR enrichment at the effector-target cell interface is more sensitive than signaling to the strength of ligand binding. Eur. J. Immunol. 33:1084–1093. [DOI] [PubMed] [Google Scholar]
- Bunnell, S.C., D.I. Hong, J.R. Kardon, T. Yamazaki, C.J. McGlade, V.A. Barr, and L.E. Samelson. 2002. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158:1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchat, A.F., D.J. Calderwood, G.C. Hirst, N.J. Holman, D.N. Johnston, R. Munschauer, P. Rafferty, and G.B. Tometzki. 2000. Pyrrolo[2,3-d]pyrimidines containing an extended 5-substituent as potent and selective inhibitors of lck II. Bioorg. Med. Chem. Lett. 10:2171–2174. [DOI] [PubMed] [Google Scholar]
- Burshtyn, D.N., A.M. Scharenberg, N. Wagtmann, S. Rajagopalan, K. Berrada, T. Yi, J.P. Kinet, and E.O. Long. 1996. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity. 4:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burshtyn, D.N., A.S. Lam, M. Weston, N. Gupta, P.A. Warmerdam, and E.O. Long. 1999. Conserved residues amino-terminal of cytoplasmic tyrosines contribute to the SHP-1-mediated inhibitory function of killer cell Ig-like receptors. J. Immunol. 162:897–902. [PubMed] [Google Scholar]
- Campi, G., R. Varma, and M.L. Dustin. 2005. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin, L.M., K. Eleme, F.E. McCann, and D.M. Davis. 2001. Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses. J. Exp. Med. 194:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, G.B., R.T. Gandhi, D.M. Davis, O. Mandelboim, B.K. Chen, J.L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 10:661–671. [DOI] [PubMed] [Google Scholar]
- Colonna, M., and J. Samaridis. 1995. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 268:405–408. [DOI] [PubMed] [Google Scholar]
- Davis, D.M., and M.L. Dustin. 2004. What is the importance of the immunological synapse? Trends Immunol. 25:323–327. [DOI] [PubMed] [Google Scholar]
- Davis, D.M., I. Chiu, M. Fassett, G.B. Cohen, O. Mandelboim, and J.L. Strominger. 1999. The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. USA. 96:15062–15067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass, A.D., and R.D. Vale. 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 121:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich, L.I., P.J. Ebert, M.F. Krummel, A. Weiss, and M.M. Davis. 2002. Dynamics of p56lck translocation to the T cell immunological synapse following agonist and antagonist stimulation. Immunity. 17:809–822. [DOI] [PubMed] [Google Scholar]
- Eleme, K., S.B. Taner, B. Onfelt, L.M. Collinson, F.E. McCann, N.J. Chalupny, D. Cosman, C. Hopkins, A.I. Magee, and D.M. Davis. 2004. Cell surface organization of stress-inducible proteins ULBP and MICA that stimulate human NK cells and T cells via NKG2D. J. Exp. Med. 199:1005–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman, D.M. 2005. Membranes are more mosaic than fluid. Nature. 438:578–580. [DOI] [PubMed] [Google Scholar]
- Eriksson, M., G. Leitz, E. Fallman, O. Axner, J.C. Ryan, M.C. Nakamura, and C.L. Sentman. 1999. Inhibitory receptors alter natural killer cell interactions with target cells yet allow simultaneous killing of susceptible targets. J. Exp. Med. 190:1005–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, Q.R., E.O. Long, and D.C. Wiley. 2000. Cobalt-mediated dimerization of the human natural killer cell inhibitory receptor. J. Biol. Chem. 275:23700–23706. [DOI] [PubMed] [Google Scholar]
- Fassett, M.S., D.M. Davis, M.M. Valter, G.B. Cohen, and J.L. Strominger. 2001. Signaling at the inhibitory natural killer cell immune synapse regulates lipid raft polarization but not class I MHC clustering. Proc. Natl. Acad. Sci. USA. 98:14547–14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, M., D.F. Barber, S.M. Takahashi, T. Jin, and E.O. Long. 2003. Spontaneous clustering and tyrosine phosphorylation of NK cell inhibitory receptor induced by ligand binding. J. Immunol. 170:6107–6114. [DOI] [PubMed] [Google Scholar]
- Fry, A.M., L.L. Lanier, and A. Weiss. 1996. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J. Exp. Med. 184:295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley, Q.S., V. Subramaniam, D.J. Arndt-Jovin, and T.M. Jovin. 2001. Fluorescence lifetime imaging: multi-point calibration, minimum resolvable differences, and artifact suppression. Cytometry. 43:248–260. [DOI] [PubMed] [Google Scholar]
- Karlhofer, F.M., R.K. Ribaudo, and W.M. Yokoyama. 1992. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 358:66–70. [DOI] [PubMed] [Google Scholar]
- Karre, K., H.G. Ljunggren, G. Piontek, and R. Kiessling. 2005. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. 1986. J. Immunol. 174:6566–6569. [PubMed] [Google Scholar]
- Krzewski, K., X. Chen, J.S. Orange, and J.L. Strominger. 2006. Formation of a WIP-, WASp-, actin-, and myosin IIA-containing multiprotein complex in activated NK cells and its alteration by KIR inhibitory signaling. J. Cell Biol. 173:121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi, A., C. Nakada, K. Ritchie, K. Murase, K. Suzuki, H. Murakoshi, R.S. Kasai, J. Kondo, and T. Fujiwara. 2005. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34:351–378. [DOI] [PubMed] [Google Scholar]
- Lanier, L.L. 2005. a. Missing self, NK cells, and The White Album. J. Immunol. 174:6565. [DOI] [PubMed] [Google Scholar]
- Lanier, L.L. 2005. b. NK cell recognition. Annu. Rev. Immunol. 23:225–274. [DOI] [PubMed] [Google Scholar]
- Ljunggren, H.G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 11:237–244. [DOI] [PubMed] [Google Scholar]
- Lou, Z., D. Jevremovic, D.D. Billadeau, and P.J. Leibson. 2000. A balance between positive and negative signals in cytotoxic lymphocytes regulates the polarization of lipid rafts during the development of cell-mediated killing. J. Exp. Med. 191:347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, T., A. Squire, G. Hansra, F. Bornancin, C. Prevostel, A. Hanby, W. Harris, D. Barnes, S. Schmidt, H. Mellor, et al. 1999. Imaging protein kinase Cα activation in cells. Science. 283:2085–2089. [DOI] [PubMed] [Google Scholar]
- Olcese, L., P. Lang, F. Vely, A. Cambiaggi, D. Marguet, M. Blery, K.L. Hippen, R. Biassoni, A. Moretta, L. Moretta, et al. 1996. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J. Immunol. 156:4531–4534. [PubMed] [Google Scholar]
- Orange, J.S., K.E. Harris, M.M. Andzelm, M.M. Valter, R.S. Geha, and J.L. Strominger. 2003. The mature activating natural killer cell immunologic synapse is formed in distinct stages. Proc. Natl. Acad. Sci. USA. 100:14151–14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan, S., C.C. Winter, N. Wagtmann, and E.O. Long. 1995. The Ig-related killer cell inhibitory receptor binds zinc and requires zinc for recognition of HLA-C on target cells. J. Immunol. 155:4143–4146. [PubMed] [Google Scholar]
- Saito, T., and T. Yokosuka. 2006. Immunological synapse and microclusters: the site for recognition and activation of T cells. Curr. Opin. Immunol. 18:305–313. [DOI] [PubMed] [Google Scholar]
- Standeven, L.J., L.M. Carlin, P. Borszcz, D.M. Davis, and D.N. Burshtyn. 2004. The actin cytoskeleton controls the efficiency of killer Ig-like receptor accumulation at inhibitory NK cell immune synapses. J. Immunol. 173:5617–5625. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.C., C. Watzl, D.D. Billadeau, P.J. Leibson, D.N. Burshtyn, and E.O. Long. 2003. Vav1 dephosphorylation by the tyrosine phosphatase SHP-1 as a mechanism for inhibition of cellular cytotoxicity. Mol. Cell. Biol. 23:6291–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhling, K., P.M. French, and D. Phillips. 2005. Time-resolved fluorescence microscopy. Photochem. Photobiol. Sci. 4:13–22. [DOI] [PubMed] [Google Scholar]
- Trautmann, A. 2005. Microclusters initiate and sustain T cell signaling. Nat. Immunol. 6:1213–1214. [DOI] [PubMed] [Google Scholar]
- Treanor, B., P.M. Lanigan, K. Suhling, T. Schreiber, I. Munro, M.A. Neil, D. Phillips, D.M. Davis, and P.M. French. 2005. Imaging fluorescence lifetime heterogeneity applied to GFP-tagged MHC protein at an immunological synapse. J. Microsc. 217:36–43. [DOI] [PubMed] [Google Scholar]
- Veillette, A., S. Latour, and D. Davidson. 2002. Negative regulation of immunoreceptor signaling. Annu. Rev. Immunol. 20:669–707. [DOI] [PubMed] [Google Scholar]
- Verveer, P.J., F.S. Wouters, A.R. Reynolds, and P.I. Bastiaens. 2000. Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science. 290:1567–1570. [DOI] [PubMed] [Google Scholar]
- Vivier, E., J.A. Nunes, and F. Vely. 2004. Natural killer cell signaling pathways. Science. 306:1517–1519. [DOI] [PubMed] [Google Scholar]
- Vyas, Y.M., K.M. Mehta, M. Morgan, H. Maniar, L. Butros, S. Jung, J.K. Burkhardt, and B. Dupont. 2001. Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J. Immunol. 167:4358–4367. [DOI] [PubMed] [Google Scholar]
- Vyas, Y.M., H. Maniar, and B. Dupont. 2002. Cutting edge: differential segregation of the SRC homology 2-containing protein tyrosine phosphatase-1 within the early NK cell immune synapse distinguishes noncytolytic from cytolytic interactions. J. Immunol. 168:3150–3154. [DOI] [PubMed] [Google Scholar]
- Wagtmann, N., S. Rajagopalan, C.C. Winter, M. Peruzzi, and E.O. Long. 1995. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 3:801–809. [DOI] [PubMed] [Google Scholar]
- Wu, P., and L. Brand. 1994. Resonance energy transfer: methods and applications. Anal. Biochem. 218:1–13. [DOI] [PubMed] [Google Scholar]
- Yokosuka, T., K. Sakata-Sogawa, W. Kobayashi, M. Hiroshima, A. Hashimoto-Tane, M. Tokunaga, M.L. Dustin, and T. Saito. 2005. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6:1253–1262. [DOI] [PubMed] [Google Scholar]
- Yokoyama, W.M., and W.E. Seaman. 1993. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu. Rev. Immunol. 11:613–635. [DOI] [PubMed] [Google Scholar]