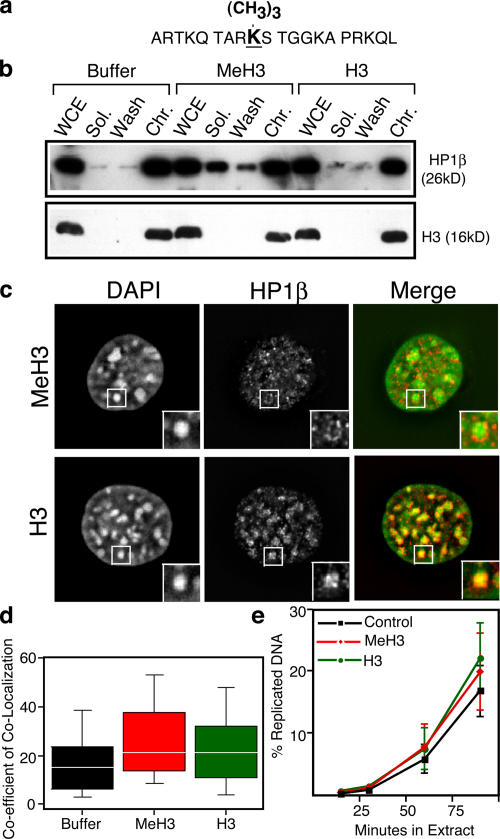
Figure 4.
Removal of HP1 proteins from chromatin does not permit early replication of chromocenters. Nuclei prepared from C127 cells synchronized at 3 h after mitosis were incubated for 2 h on ice in the presence of 50 μg/ml of either methylated or unmethylated peptide or buffer alone. (a) Peptide used for HP1 competition. The underlined amino acid is the attachment site for the methyl group. (b) Aliquots of nuclei were extracted, and fractions were analyzed by immunoblotting as in Fig. 3 (0.15 M NaCl). Adding additional methylated peptide or a combination of chromoshadow peptides (Smothers and Henikoff, 2000) and methyl-H3 peptides did not remove additional HP1 proteins (not depicted). WCE, whole cell extract. (c) Aliquots of the same nuclei were stained with anti-HP1 antibody (red) and counterstained with DAPI (pseudocolored green). The insets show higher magnification images of one chromocenter each. HP1 was not detected in the core of the chromocenters after extraction with methylated peptide, although the less DAPI-dense chromatin surrounding the chromocenters retained some HP1. (d) Additional aliquots of the same nuclei were introduced into Xenopus egg extract, and the replication timing of chromocenters was evaluated as in Fig. 1 except that the analysis was restricted to the 30-min time point. Shown is a box plot (displayed as in Fig. 1) for >200 nuclei from three independent experiments (40–80 nuclei/experiment). (e) Aliquots of the Xenopus egg extract reaction were supplemented with α-[32P]dATP, and the percentage of the total input genomic DNA synthesized was evaluated at each of the indicated time points as in Fig. 1. The means and SEM (error bars) for three independent experiments are shown.