Abstract
Previous evidence has indicated that an intact centrosome is essential for cell cycle progress and that elimination of the centrosome or depletion of individual centrosome proteins prevents the entry into S phase. To investigate the molecular mechanisms of centrosome-dependent cell cycle progress, we performed RNA silencing experiments of two centrosome-associated proteins, pericentriolar material 1 (PCM-1) and pericentrin, in primary human fibroblasts. We found that cells depleted of PCM-1 or pericentrin show lower levels of markers for S phase and cell proliferation, including cyclin A, Ki-67, proliferating cell nuclear antigen, minichromosome maintenance deficient 3, and phosphorylated retinoblastoma protein. Also, the percentage of cells undergoing DNA replication was reduced by >50%. At the same time, levels of p53 and p21 increased in these cells, and cells were predisposed to undergo senescence. Conversely, depletion of centrosome proteins in cells lacking p53 did not cause any cell cycle arrest. Inhibition of p38 mitogen-activated protein kinase rescued cell cycle activity after centrosome protein depletion, indicating that p53 is activated by the p38 stress pathway.
Introduction
The centrosome nucleates and organizes microtubules in animal cells. It consists of a pair of cylindrically shaped centrioles surrounded by fibrous pericentriolar material. Before or during DNA replication in S phase, the centrioles split, and each cylinder serves as a template for the assembly of a new “daughter” centriole. Before mitosis, when cells contain two pairs of centrioles, each pair serves as a nucleation center for microtubules of the spindle apparatus. Defects in centrosome assembly or in centrosome separation can result in defective nucleation of spindle microtubules, and in several cases, in the formation of monopolar spindles and mitotic arrest (Sunkel et al., 1995; Faragher and Fry, 2003). Several years ago, evidence was published that defective centrosome assembly can prevent cells from entering S phase. In particular, removal of the centrosome by microsurgery or by laser ablation resulted in a cell cycle arrest, as did inhibition or silencing of several centrosome-associated proteins, such as dynactin, PARP-3, centriolin, or AKAP450 (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001; Quintyne and Schroer, 2002; Augustin et al., 2003; Gromley et al., 2003; Keryer et al., 2003). The mechanism leading to this centrosome-dependent cell cycle arrest in G1 phase has been unclear; it was proposed that a checkpoint control would prevent those cells with imperfect centrosomes from continuing the cell cycle, to prevent the assembly of defective spindles later in mitosis (Murray, 2001).
In this study, we followed cell cycle progress after inhibition of centrosome assembly by depleting the pericentriolar proteins pericentriolar material 1 (PCM-1) and pericentrin. These proteins have been shown to be necessary for the assembly of other centrosomal constituents (Dictenberg et al., 1998; Dammermann and Merdes, 2002; Kubo and Tsukita, 2003). We found that depletion of PCM-1 or pericentrin activates the p38-dependent stress pathway and the p53-dependent cell cycle checkpoint.
Results and discussion
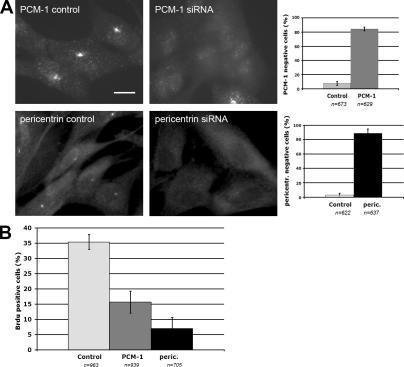
We have previously shown that depletion of the protein PCM-1 leads to defects in the assembly of the centrosomal components centrin, ninein, and pericentrin, and to an altered organization of the microtubule network in interphase cells (Dammermann and Merdes, 2002). To investigate the consequences of PCM-1 depletion on the cell cycle, we performed RNA silencing experiments in primary human fibroblasts, MRC-5. After 72 h, PCM-1 depletion was monitored by immunofluorescence (Fig. 1 A) and Western blotting (Fig. 2 A). Depleted cells were tested for incorporation of BrdU into the nucleus, as an indicator of DNA synthesis (Fig. 1 B). We determined that in PCM-1–depleted cells only 15 ± 4% incorporated BrdU, as compared with 35 ± 3% in controls, as expected for a normal cycling population (Fig. 1 B). This is consistent with previous reports on microinjection of PCM-1–inhibiting antibodies (Balczon et al., 2002) and on centrosome removal by microsurgery or laser ablation, which prevent cells from entering S phase (Hinchcliffe et al., 2001; Khodjakov and Rieder, 2001). Several years ago, experiments on cells treated with the microtubule drugs colcemid, nocodazole, and taxol indicated that untransformed cells are arrested in G1 phase, when microtubules are depolymerized or when microtubule dynamics are altered (Trielli et al., 1996; Di Leonardo et al., 1997; Lanni and Jacks, 1998). This raises the question of whether DNA replication in PCM-1–depleted cells is inhibited because of an altered microtubule network, or whether defects at the centrosome itself suffice to induce a cell cycle arrest. Therefore, we depleted a second centrosome protein, pericentrin (Fig. 1 A), which in contrast to PCM-1, only slightly reduces microtubule density but seems to have no significant effect on microtubule anchoring at the centrosome (Dammermann and Merdes, 2002). Consistently, depletion of pericentrin also led to a reduction of BrdU incorporation (Fig. 1 B).
Figure 1.
Depletion of PCM-1 and pericentrin prevents DNA replication. (A) Immunofluorescence of MRC-5 fibroblasts treated with control RNA or siRNA against PCM-1 and pericentrin, respectively. Bar, 10 μm. Graphs depict the percentages of cells lacking PCM-1 or pericentrin expression after treatment with control or silencing RNAs. Cells were scored as negative if they lacked centrosomal staining of PCM-1 or pericentrin, only displaying diffuse background fluorescence. (B) Percentages of cells incorporating BrdU after treatment with control RNA or silencing RNA against PCM-1 or pericentrin. Data were obtained from cell cultures that were double labeled for BrdU and PCM-1 or pericentrin. Among cell cultures treated with silencing RNA, only cells that were visibly depleted of PCM-1 or pericentrin were scored. Error bars indicate SD.
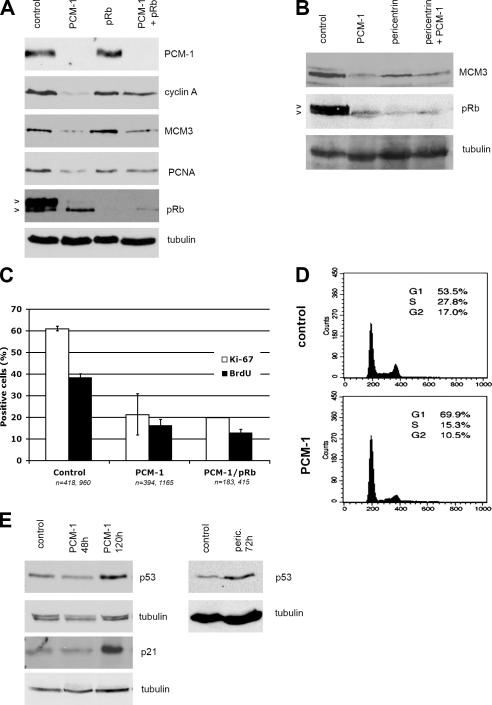
Figure 2.
Depletion of PCM-1 leads to cell cycle arrest in G1 or G0 phase. (A) Immunoblots of MRC-5 cells treated with control RNA or siRNA against PCM-1, pRb, or both PCM-1 and pRb simultaneously. Blots were probed with antibodies against PCM-1, cyclin A, the licensing factor MCM3, the DNA replication factor PCNA, pRb, and α-tubulin. (B) Immunoblots of cells treated with control RNA or siRNAs against PCM-1, pericentrin, or both simultaneously. Blots were probed with antibodies against MCM3, pRb, and α-tubulin. Arrowheads in A and B indicate migration positions of hyper- and hypophosphorylated pRb. (C) MRC-5 cells treated with control RNA, siRNA against PCM-1, or siRNA against PCM-1 and pRb simultaneously. The percentages of cells staining positively for the proliferation marker Ki-67 or for incorporated BrdU are shown. Experimental conditions were similar to Fig. 1 B. Error bars indicate SD. (D) Flow-cytometric analysis of MRC-5 cells treated with control RNA or siRNA against PCM-1. (E) Immunoblots of MRC-5 cells treated with control RNA or siRNA against PCM-1 for 48 or 120 h or with siRNA against pericentrin for 72 h. Blots were probed with antibodies against p53, α-tubulin, or p21.
Because these data indicated that cells failed to undergo S phase–dependent DNA replication when missing the full complement of centrosome proteins, we wanted to test in more detail at what stage cells are arrested. Immunoblotting of cell extracts of PCM-1–depleted cultures indicated drastic reduction of cyclin A (Fig. 2 A), which is normally found expressed in cells during late G1, S, and G2 phases. Furthermore, we detected that PCM-1–depleted cells also showed reduced amounts of the proteins minichromosome maintenance deficient 3 (MCM3) and proliferating cell nuclear antigen (PCNA) acting in licensing and DNA replication (Fig. 2 A) and reduced percentages of cells expressing the cell proliferation marker Ki-67 (Fig. 2 C). Equivalent data were also obtained by depletion of pericentrin (Fig. 2 B). Altogether, these data indicated that depletion of centrosome proteins reduces the number of cells entering S phase. We verified this hypothesis by comparing profiles of cell cultures analyzed by flow cytometry (Fig. 2 D): consistent with our data on BrdU incorporation, PCM-1–depleted cells showed a reduction of S phases from 28 to 15%.
We then wanted to determine whether S phase entry was blocked because of checkpoint activation in cells depleted of PCM-1 or pericentrin. We found that the overall levels of the retinoblastoma protein (pRb) were reduced to 38 ± 17% and that most of the remaining pRb, normally hyperphosphorylated during late G1 and S phase, was present in its faster migrating, hypophosphorylated form (77 ± 13% in depleted vs. 38 ± 13% in control cells; Fig. 2, A and B). The checkpoint protein p53, however, was found up-regulated, especially after prolonged depletion of PCM-1 or pericentrin (Fig. 2 E). In depleted cells, the Cdk2 inhibitor p21 was found equally up-regulated (Fig. 2 E). These data suggested that depletion of the two centrosome-associated proteins PCM-1 and pericentrin leads to the activation of the p53-dependent checkpoint.
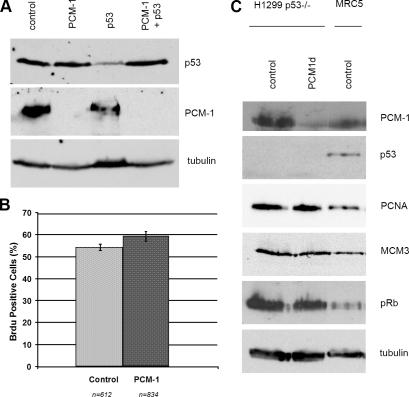
In the next step, we wanted to determine whether cell cycle progress would be affected if the p53-dependent checkpoint control was abrogated. For this purpose, we attempted simultaneous depletion of p53 and PCM-1 by cotransfecting siRNA oligomers against both. Fig. 3 A shows that p53 levels could not be reduced when PCM-1 was missing. We tried to refine this experiment by sequential depletion of MRC-5 cells, by first depleting >90% of p53 after 72 h, followed by simultaneous siRNA treatment against p53 and PCM-1. However, we observed that under these conditions, p53 levels increased back to 40–50% in three different experiments (unpublished data). We concluded that p53 turnover is altered and that the residual p53 protein might be stabilized in the absence of an intact centrosome. We therefore changed our experimental strategy and compared cell cycle progress after PCM-1 depletion in several cell lines lacking p53. We used mouse embryonic fibroblasts from p53 knockout mice (unpublished data) as well as the human lung carcinoma cell line H1299. Because of the loss of p53 checkpoint control, both lines displayed a relatively high basic rate of DNA synthesis (Fig. 3 B). We found that in both p53−/− cell lines, PCM-1 depletion did not inhibit cell cycle progress. The levels of PCNA, MCM3, and hyperphosphorylated pRb remained high (Fig. 3, B and C, H1299). In addition, we also tested the effect of PCM-1 depletion in the p53+/+ and p53−/− lines of HCT116 cells. Unfortunately, only 30% of these cells showed lower PCM-1 levels. BrdU incorporation in these was reduced from 42% in controls to 33% in partially depleted p53+ cells, whereas p53− cells showed BrdU incorporation in 49% after partial PCM-1 depletion. Consistently, HeLa cells with functionally suppressed p53 checkpoint control do not arrest in the absence of centrosomes (La Terra et al., 2005). In contrast to p53, the removal of pRb did not cause resumption of the cell cycle in PCM-1–depleted MRC-5 cells, because the percentages of BrdU-incorporating cells and Ki-67–expressing cells remained low, as did the expression of MCM3 protein (Fig. 2, A and C). On the other hand, the amounts of cyclin A and PCNA were restored to nearly control levels. A possible scenario would be that centrosome defects activate both pRb and p53 in parallel, with p53 having feedback effects on pRb phosphorylation and pRb protein levels but not vice versa. This would be consistent with our observation that pRb levels drop after PCM-1 or pericentrin depletion and that depletion of pRb itself does not fully restore S phase activity, which is probably blocked because of checkpoint control mechanisms directly dependent on p53. Eventually, loss of pRb might be compensated by Rb-related “pocket proteins,” such as p107 or p130.
Figure 3.
Cell cycle arrest after PCM-1 depletion depends on the checkpoint protein p53. (A) Immunoblots of MRC-5 cells treated with control RNA or siRNAs against PCM-1, p53, or both PCM-1 and p53 combined. Blots were probed with antibodies against p53, PCM-1, or α-tubulin. (B) Graph depicts BrdU incorporation in p53−/− H1299 cells treated with control RNA or siRNA against PCM-1. Error bars indicate SD. (C) Immunoblots of H1299 and MRC-5 cells treated with control RNA or siRNA against PCM-1 (PCM1d) for 72 h. Blots were probed with antibodies against PCM-1, p53, PCNA, MCM3, pRb, or α-tubulin.
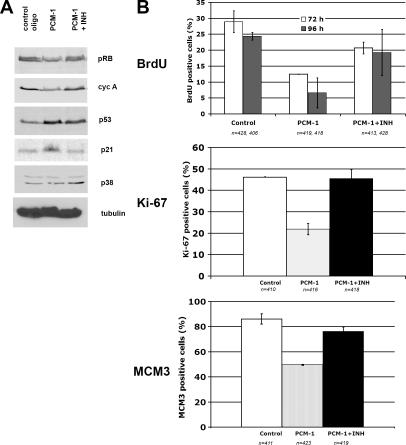
Finally, we addressed the question of how defects in centrosome assembly could activate the p53-dependent checkpoint. It has been reported that p38 MAPK is involved in the cell's response to a range of stress factors, such as UV irradiation, osmotic shock, heat shock, starvation, and cytokine treatment (Zarubin and Han, 2005). The response to these stress factors is mediated partly through p53 phosphorylation, which is believed to stabilize p53 and to increase the expression of the Cdk inhibitor p21, thereby blocking the cell cycle (Agarwal et al., 1998). To assess the involvement of stress-activated p38 MAPK in response to centrosome defects, we have treated PCM-depleted MRC-5 cells with the p38 MAPK–specific inhibitor SB203580. This inhibitor blocks p38 activity but affects neither p38 protein levels nor p38 phosphorylation. We found that the inhibitor prevented cell cycle arrest, as indicated by resumption of DNA synthesis (Fig. 4 B). p38 inhibition in PCM-1–depleted cells also restored regular levels of cyclin A and hyperphosphorylated pRb, as well as expression of Ki-67 and MCM3 (Fig. 4). Further, the inhibition of p38 led to decreased levels of p21 but only slightly decreased p53 (Fig. 4 A). This could be explained by a stabilization of p53 after centrosome inactivation, as discussed in the previous paragraph, which might occur independently of p38-dependent p53 activation. Alternatively, the centrosome-dependent cell cycle arrest might not simply be a linear consequence of p53 activation via p38. For example, p53 might be activated by other kinases in addition to p38. Moreover, although p38 has been shown in multiple experiments to mediate cell cycle arrest by phosphorylating various sites of p53 (Bulavin et al., 1999; Huang et al., 1999), p38 is also known to phosphorylate and thereby inactivate Cdc25A and cyclin D, which in turn arrests the cell cycle (Lavoie et al., 1996; Casanovas et al., 2000; Goloudina et al., 2003; Khaled et al., 2005). However, p38 MAPK activation in the absence of p53 seems to be insufficient for centrosome-dependent cell cycle arrest because PCM-1 depletion in p53−/− cells did not stop the cell cycle.
Figure 4.
Inhibition of the stress kinase p38 MAPK prevents cell cycle arrest after PCM-1 depletion. (A) Immunoblots of MRC-5 cells treated with control RNA, siRNA against PCM-1, or siRNA against PCM-1 together with the p38 inhibitor SB203580 (PCM-1 + INH). Blots were probed with antibodies against pRb, cyclin A (cyc A), p53, the Cdk inhibitor p21, active p38, or α-tubulin. (B) Graphs depicting MRC-5 cells treated with control RNA, siRNA against PCM-1, or siRNA against PCM-1 together with the p38 inhibitor SB203580 (PCM-1 + INH). The staining of BrdU incorporation (after RNA treatment for 72 or 96 h), Ki-67 immunofluorescence (after RNA treatment for 96 h), or MCM3 immunofluorescence (after RNA treatment for 72 h) is shown.
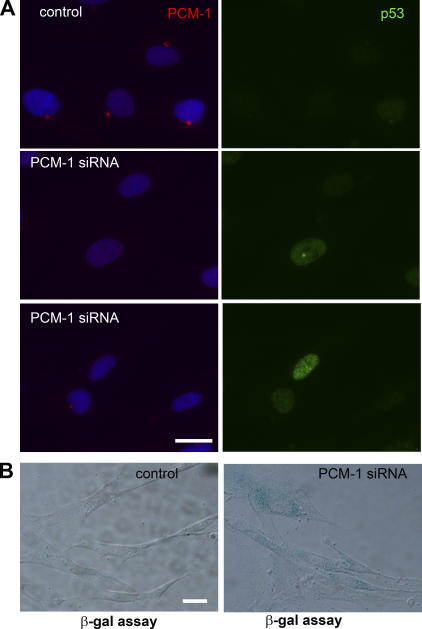
In our PCM-1–depletion experiments, we observed a significant decrease in the expression of proteins associated with cell proliferation, such as MCM3, PCNA, or Ki67. However, because about half of the depleted cells were still entering S phase, we wanted to test whether centrosome defects only slowed down cell cycle activity, or whether depleted cells were predisposed to a stable arrest that did not affect all cells simultaneously. Immunofluorescence of p53 after PCM-1 depletion revealed that p53 levels varied between individual cells, with some cells showing a very high increase but a small increase or no obvious response in others (Fig. 5 A). We then tested whether the cells' response to centrosome inhibition induces exit from the cell cycle via senescence. Senescence is marked by the acquisition of a permanent G0 state and an increase of cellular β-galactosidase activity (Dimri et al., 1995). A cytochemical analysis of PCM-1–depleted cultures revealed a 10-fold increase of β-galactosidase–positive cells (Fig. 5 B), compared with a basal level of 3% in controls. Consistently, we previously reported that cultures of U2OS cells died after silencing of PCM-1 for long periods (Dammermann and Merdes, 2002), although the experimental conditions varied from the protocol used here. Senescence has been characterized as a cellular program activated as a result of physiological stresses preventing further cell proliferation (Ben-Porath and Weinberg, 2004). Instead of a specific centrosome-dependent cell cycle control as proposed by Murray (2001), we propose a stress-driven response to centrosome defects. The idea of the centrosome as a center for stress-related signaling seems plausible, considering its central location in the focus of the microtubule network and its ability to bind various molecules of signaling pathways and of cell cycle regulation. For example, significant amounts of protein kinase A, polo-like kinase, and protein phosphatases 1 and 2A, as well as cyclin E and p53, have all been localized to the centrosome (Fry et al., 2000; Morris et al., 2000; Matsumoto and Maller, 2004). Their localization and anchoring is mediated by large coiled-coil proteins such as pericentrin, AKAP450, or AKAP220 (Diviani et al., 2000; Reinton et al., 2000; Diviani and Scott, 2001). Absence or inhibition of anchoring proteins could therefore disrupt cellular signaling pathways and elicit a stress response. Further research will be necessary to investigate the multiple interactions between centrosome proteins, cell signaling, and the complex regulation of the cell cycle.
Figure 5.
PCM-1–depleted cells are predisposed to exit from the cell cycle and to undergo senescence. (A) Fibroblasts treated with control and PCM-1 siRNA, stained for PCM-1 (red), DNA (blue), and p53 (green). (B) β-Galactosidase assay on control and PCM-1–depleted cells 96 h after transfection using 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside as a chromogenic substrate. Bars, 10 μm.
Materials and methods
Cell culture and transfection experiments
Human fetal lung fibroblasts MRC-5 (European Collection of Cell Cultures) and H1299 cells (a gift from B. Vojtesek, Masaryk Memorial Cancer Institute, Brno, Czech Republic) were cultured in DME (Invitrogen), supplemented with 10% fetal bovine serum (Perbio Science), 2 mM l-glutamine, and antibiotics. HCT116 cells (a gift from B. Vogelstein, Johns Hopkins University, Baltimore, MD) were cultured in McCoy's medium containing the same supplements. The cells were plated to reach 80% confluency at the time of transfection. About 3 × 106 cells from early passages of MRC-5 and 0.5 × 106 cells of H1299 or HCT116 were transfected with 9 or 18 μg of siRNA oligonucleotides, respectively, in a Nucleofector electroporation device, using the appropriate transfection kit (Amaxa). The cells were then plated onto two 10-cm Petri dishes to obtain a confluency of ∼30–40% on the next day. Routinely, siRNA treatment was for a total of 72 h before fixation or preparation of cell extracts, unless indicated otherwise. P38 MAPK inhibition was performed in cells that were transfected with oligos and cultured overnight before the specific inhibitor SB203580 (Calbiochem) was added at a concentration of 10 μM. Inhibitor-treated cells were left in culture for another 48 h and then fixed in methanol or extracted for SDS-PAGE.
Antibodies and siRNA oligonucleotides
Cells were harvested using trypsin-EDTA, counted, and washed in ice-cold PBS. Cells were extracted in SDS-PAGE sample buffer and boiled for 6 min. Amounts of extract equal to 200,000 cells were loaded and run on 7.5, 10, or 12.5% SDS-PAGE gels and blotted onto nitrocellulose. The following antibodies were used for Western blot analysis or immunofluorescence: affinity-purified rabbit anti–PCM-1 (Dammermann and Merdes, 2002), anti-MCM3 mAb clone 3A2 (MBL International Corporation), anti-pRB mAb clone 4H1 (Cell Signaling), anti-PCNA mAb clone PC-10 (Sigma-Aldrich), anti–cyclin E mAb clone HE12 (Zymed Laboratories), anti–cyclin A mAb clone Cy-A1 (Sigma-Aldrich), anti p53 mAb clone DO-1 (Novocastra), anti-p21 mAb clone SX118 (BD Biosciences), rabbit anti-ACTIVEp38MAPK polyclonal antibody (Promega), anti–α-tubulin mAb clone DM1A (Sigma-Aldrich), goat anti–mouse polyclonal antibody HRP (Promega), donkey anti–rabbit IgG polyclonal antibody HRP (GE Healthcare), rat anti-BrdU mAb (Harlan), anti–Ki-67 mAb clone MM1 (Novocastra), rabbit anti-pericentrin polyclonal antibody (Covance), donkey anti–rabbit or anti–mouse IgG conjugated with Alexa 488 or Alexa 594 (Invitrogen). Quantification of protein levels was performed by scanning immunoblots and analyzed using the Photoshop (Adobe) histogram tool. Quantification of cells expressing specific proteins was performed by counting siRNA-treated cells that were double stained with PCM-1 antibodies to verify depletion. The following siRNA oligomers with dTdT overhangs (QIAGEN) were used: PCM-1.2, corresponding to human PCM-1 (UCAGCUUCGUGAUUCUCAG); peric, corresponding to human pericentrin (GCAGCUGAGCUGAAGGAGA; Dammermann and Merdes, 2002); pRB (GCCCUUACAAGUUUCCUAG); and p53 (CUACUUCCUGAAAACAACG).
Flow cytometry analysis
All cells in a culture dish were harvested by trypsinization, washed in ice-cold PBS, and fixed in 80% ice-cold ethanol in PBS. Before staining, the cells were spun down in a cooled centrifuge and resuspended in the cold. Bovine pancreatic RNase (Sigma-Aldrich) was added at a final concentration of 2 μg/ml, and cells were incubated at 37°C for 30 min, followed by an incubation in 20 μg/ml of propidium iodide (Sigma-Aldrich) for 20 min at room temperature. 10,000 cells were analyzed on a flow cytometer (FACSCalibur; BD Biosciences).
β-Galactosidase assay
β-Galactosidase staining at pH 6.0 was performed as described in Dimri et al. (1995).
Immunofluorescence microscopy
Cells grown on coverslips were fixed in ice-cold methanol and stored at −20°C until use. Antibody staining was performed using the reagents listed in the previous paragraphs, according to standard protocols. The percentage of cells in S phase was assessed by adding 100 μM BrdU (Sigma-Aldrich) to the cultures 30 min before fixation. Double labeling of BrdU and PCM-1 was performed by probing first for PCM-1, using rabbit anti–PCM-1 and fluorescent anti-rabbit antibody, and coverslips were postfixed in PBS containing 3.7% paraformaldehyde, treated with 2 M HCl for 30 min, and stained with rat anti-BrdU and fluorescent secondary anti-rat antibody. Cells were viewed with a fluorescence microscope (Axioskop 2; Carl Zeiss MicroImaging, Inc.) equipped with a camera (Axiocam; Carl Zeiss MicroImaging, Inc.) and software (Axiovision; Carl Zeiss MicroImaging, Inc.). The images were imported into Photoshop for presentation.
Acknowledgments
We thank our colleagues at the University of Edinburgh, in particular, Dr. W.C. Earnshaw and members of his group for their help and for stimulating discussions, Dr. B. Vogelstein for HCT116 cells, and Dr. B. Vojtesek for H1299 cells.
This work was supported by a Wellcome Trust Senior Research Fellowship to A. Merdes.
A. Dammermann's present address is Ludwig Institute for Cancer Research, University of California, San Diego, La Jolla, CA 92093.
A. Merdes' present address is Centre National de la Recherche Scientifique–Pierre Fabre, 31400 Toulouse, France.
Abbreviations used in this paper: MCM3, minichromosome maintenance deficient 3; PCM-1, pericentriolar material 1; PCNA, proliferating cell nuclear antigen; pRb, retinoblastoma protein.
References
- Agarwal, M.L., W.R. Taylor, M.V. Chernov, O.B. Chernova, and G.R. Stark. 1998. The p53 network. J. Biol. Chem. 273:1–4. [DOI] [PubMed] [Google Scholar]
- Augustin, A., C. Spenlehauer, H. Dumond, J. Menissier-De Murcia, M. Piel, A.C. Schmit, F. Apiou, J.L. Vonesch, M. Kock, M. Bornens, and G. De Murcia. 2003. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J. Cell Sci. 116:1551–1562. [DOI] [PubMed] [Google Scholar]
- Balczon, R., C. Simerly, D. Takahashi, and G. Schatten. 2002. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil. Cytoskeleton. 52:183–192. [DOI] [PubMed] [Google Scholar]
- Ben-Porath, I., and R.A. Weinberg. 2004. When cells get stressed: an integrative view of cellular senescence. J. Clin. Invest. 113:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulavin, D.V., S. Saito, M.C. Hollander, K. Sakaguchi, C.W. Anderson, E. Appella, and A.J. Jr. Fornace. 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18:6845–6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanovas, O., F. Miro, J.M. Estanyol, E. Itarte, N. Agell, and O. Bachs. 2000. Osmotic stress regulates the stability of cyclin D1 in a p38SAPK2-dependent manner. J. Biol. Chem. 275:35091–35097. [DOI] [PubMed] [Google Scholar]
- Dammermann, A., and A. Merdes. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo, A., S.H. Khan, S.P. Linke, V. Greco, G. Seidita, and G.M. Wahl. 1997. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 57:1013–1019. [PubMed] [Google Scholar]
- Dictenberg, J.B., W. Zimmerman, C.A. Sparks, A. Young, C. Vidair, Y. Zheng, W. Carrington, F.S. Fay, and S.J. Doxsey. 1998. Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri, G.P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E.E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 92:9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviani, D., and J.D. Scott. 2001. AKAP signaling complexes at the cytoskeleton. J. Cell Sci. 114:1431–1437. [DOI] [PubMed] [Google Scholar]
- Diviani, D., L.K. Langeberg, S.J. Doxsey, and J.D. Scott. 2000. Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol. 10:417–420. [DOI] [PubMed] [Google Scholar]
- Faragher, A.J., and A.M. Fry. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell. 14:2876–2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, A.M., T. Mayor, and E.A. Nigg. 2000. Regulating centrosomes by protein phosphorylation. Curr. Top. Dev. Biol. 49:291–312. [DOI] [PubMed] [Google Scholar]
- Goloudina, A., H. Yamaguchi, D.B. Cheryakova, E. Appella, A.J. Fornace Jr., and D.V. Bulavin. 2003. Regulation of human Cdc25A stability by Serine 75 phosphorylation is not sufficient to activate a S-phase checkpoint. Cell Cycle. 2:473–478. [PubMed] [Google Scholar]
- Gromley, A., A. Jurczyk, J. Silibourne, F. Halilovic, M. Mogensen, I. Groisman, M. Blomberg, and S. Doxsey. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe, E.H., F.J. Miller, M. Cham, A. Khodjakov, and G. Sluder. 2001. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 291:1547–1550. [DOI] [PubMed] [Google Scholar]
- Huang, C., W.Y. Ma, A. Maxiner, Y. Sun, and Z. Dong. 1999. p38 kinase mediates UV-induced phosphorylation of p53 protein at serine 389. J. Biol. Chem. 274:12229–12235. [DOI] [PubMed] [Google Scholar]
- Keryer, G., O. Witczak, A. Delouve, W.A. Kemmner, D. Rouillard, K. Tasken, and M. Bornens. 2003. Dissociating the centrosomal matrix protein AKAP450 from centrioles impairs centriole duplication and cell cycle progression. Mol. Biol. Cell. 14:2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled, A.R., D.V. Bulavin, C. Kittipatarin, W.Q. Li, M. Alvarez, K. Kim, H.A. Young, A.J. Fornace, and S.K. Durum. 2005. Cytokine-driven cell cycling is mediated through Cdc25A. J. Cell Biol. 169:755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 2001. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J. Cell Biol. 153:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., and S. Tsukita. 2003. Non-membranous granular organelle consisting of PCM-1: subcellular distribution and cell-cycle-dependent assembly/disassembly. J. Cell Sci. 116:919–928. [DOI] [PubMed] [Google Scholar]
- Lanni, J.S., and T. Jacks. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18:1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra, S., C.N. English, P. Hergert, B.F. McEwen, G. Sluder, and A. Khodjakov. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie, J.N., G. L'Allemain, A. Brunet, R. Muller, and J. Pouyssegur. 1996. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J. Biol. Chem. 271:20608–20616. [DOI] [PubMed] [Google Scholar]
- Matsumoto, Y., and J.L. Maller. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 306:885–888. [DOI] [PubMed] [Google Scholar]
- Morris, V.B., J. Brammall, J. Noble, and R. Reddel. 2000. p53 localizes to the centrosomes and spindles of mitotic cells in the embryonic chick epiblast, human cell lines, and a human primary culture: an immunofluorescence study. Exp. Cell Res. 256:122–130. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 2001. Cell cycle. Centrioles at the checkpoint. Science. 291:1499–1502. [DOI] [PubMed] [Google Scholar]
- Quintyne, N.J., and T.A. Schroer. 2002. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159:245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinton, N., P. Collas, T.B. Haugen, B.S. Skalhegg, V. Hansson, T. Jahnsen, and K. Tasken. 2000. Localization of a novel human A-kinase-anchoring protein, hAKAP220, during spermatogenesis. Dev. Biol. 223:194–204. [DOI] [PubMed] [Google Scholar]
- Sunkel, C.E., R. Gomes, P. Sampaio, J. Perdiago, and C. Gonzalez. 1995. Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 14:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trielli, M.O., P.R. Andreassen, F.B. Lacroix, and R.B. Margolis. 1996. Differential Taxol-dependent arrest of transformed and nontransformed cells in the G1 phase of the cell cycle, and specific-related mortality of transformed cells. J. Cell Biol. 135:689–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarubin, T., and J. Han. 2005. Activation and signalling of the p38 MAP kinase pathway. Cell Res. 15:11–18. [DOI] [PubMed] [Google Scholar]