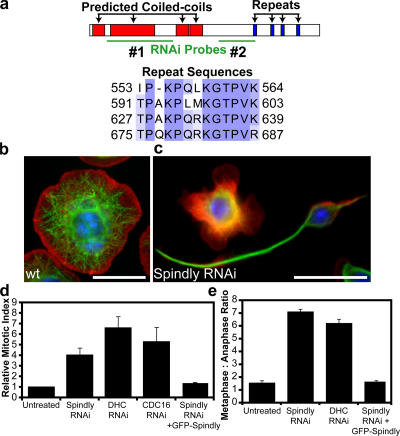
Figure 1.
RNAi of Spindly alters cell morphology and causes mitotic arrest in Drosophila S2 cells. (a) Domains of the Spindly protein showing predicted coiled-coil sequences in red and repeated motifs in blue; sequence alignment of residues in the repeat motifs is shown below. The locations of two nonoverlapping dsRNAs used to deplete Spindly are shown in green. A third dsRNA to the 3′ UTR was also used (not depicted). (b and c) Wild-type (wt) S2 cells show a uniformly spread morphology (b), whereas Spindly RNAi-treated cells (c) show marked defects in the actin lamellae as well as increased numbers of cells with long microtubule-rich projections. Actin, red; microtubules, green; DNA, blue. (d) The mitotic index of S2 cells is increased after the depletion of Spindly, dynein heavy chain (DHC), or a subunit of the APC (Cdc16; mean ± SEM [error bars]; n = 3 experiments, with 1,000–3,000 cells counted per experiment). Values are expressed as a ratio of RNAi-treated to untreated cells (untreated cells have a mitotic index of 1–3%). (e) The ratio of metaphase to anaphase cells (scored manually after staining with anti-tubulin and antiphosphohistone antibodies; see Materials and methods) reveals a selective increase in metaphase cells after Spindly and DHC RNAi (mean ± SEM; n = 2 experiments, with >200 mitotic spindles scored per experiment). (d and e) The expression of GFP-tagged Spindly can rescue the mitotic phenotype after endogenous Spindly is depleted using a dsRNA that targets the Spindly 3′ UTR. Bars, 10 μm.