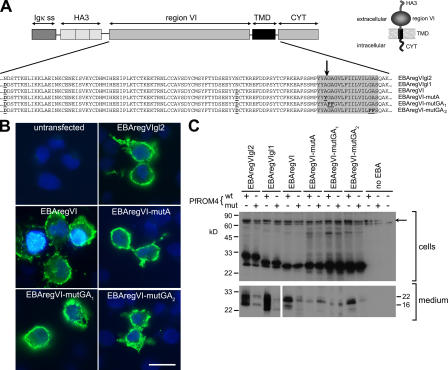
Figure 5.
EBA-175 is a substrate for PfROM4. (A) Structure of EBA-175 minigenes expressed in COS-7 cells. Partial amino acid sequences of the various mutants are indicated. Substitutions made to remove N-glycosylation sites or to modify the TMD (shaded) are in bold and underlined. The position mapped as the site of EBA-175 shedding is indicated by an arrow. The predicted topology of the expression products is shown (inset). (B) IFA showing expression of EBA-175 minigene products at the surface of COS-7 cells. Cells transfected with the various constructs were probed without fixation or permeabilization with the anti-HA mAb 3F10 (green). In all cases, 15–20% of cells exhibited strong surface fluorescence. Nuclei were stained with DAPI (blue). Bar, 50 μm. (C) Western blots of cells or medium from COS-7 cells cotransfected with EBA-175 minigene constructs and HA-tagged PfROM4 expression constructs, either in wild-type (wt) or active site Ser knockout form (mut). Blots were probed with mAb 3F10. The uppermost band visible in the cell extract blot (arrow) corresponds to HA-tagged PfROM4; note its absence from the farthest right lane, where no PfROM4 construct was transfected. The band just below this is a nonspecific reaction signal. These results were reproducible in >19 independent experiments.