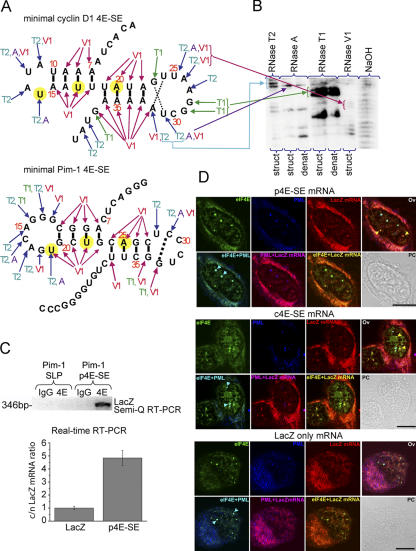
Figure 2.
A common secondary structure for the 4E-SE that acts as a zip code for eIF4E nuclear bodies. (A) Secondary structure for cyclin D1 4E-SE (c4E-SE) and Pim-1 4E-SE (p4E-SE) as determined by RNase mapping experiments. Conserved set of A and U nucleotides (UX2UX2A) are highlighted in yellow. (B) A sample gel. (C, top) Only one of the two predicted SLPs from Pim-1 3′ UTR (p4E-SE) immunoprecipitates with eIF4E. (bottom) eIF4E promotes the export of lacZ mRNA that contains minimal p4E-SE. Cytoplasmic/nuclear (c/n) values represent relative fold ± SD (error bars) normalized to the lacZ control, which was set to 1. Calculations were performed as described in Table II. (D) Colocalization of lacZ–p4E-SE, lacZ–c4E-SE, or lacZ transcripts with PML and eIF4E protein was examined in U2OS cells transfected with lacZ/lacZ–4E-SE. LacZ mRNA was detected using in situ hybridization with a biotin-labeled nick-translated probe to lacZ (red). Cells were then immunostained using an eIF4E mAb conjugated directly to FITC (green) and PML mAb 5E10 (blue). Importantly, lacZ mRNAs containing the 4E-SE from either cyclin D1 or Pim-1 colocalize to eIF4E nuclear bodies (arrowheads). As we have shown previously for endogenous cyclin D1 mRNA, there are two populations of eIF4E nuclear bodies: those that colocalize with lacZ mRNA and those that colocalize with PML. Magnification was 100× and 3× (for lacZ and c4ESE) or 4× (p4ESE) digital zoom. Bars, 10 μM.