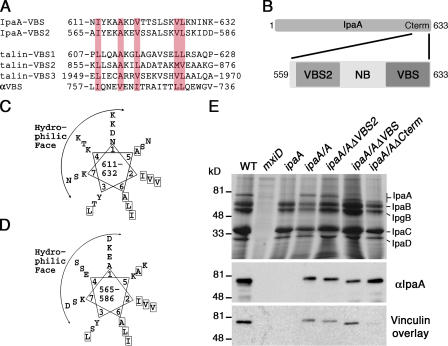
Figure 1.
S. flexneri IpaA harbors two putative VBSs (IpaA-VBSs). (A) Structure-based sequence alignment of the complete IpaA amino acid sequence, using the Heidelberg PHD secondary-structure program, predicted that IpaA contains 66% α-helical character and identified two short regions, residues 563–589 and 612–630, which share strong similarity to the VBSs of talin and α-actinin. These putative IpaA VBSs, IpaA-VBS and -VBS2, were aligned with three of the VBSs of human talin (talin-VBS1, -VBS2, and -VBS3) and that of α-actinin (αVBS) using CLUSTAL protein software. As seen in the Vh1–αVBS structure (Bois et al., 2005), the orientation of the αVBS is inverted. Hydrophobic residues conserved in all five VBSs are shaded in pink. (B) Schematic of IpaA is shown and the C-terminal region of IpaA (residues 559–633), indicating the position and nomenclature of the peptides used in this study. (C) IpaA-VBS and (D) -VBS2 are predicted to be amphipathic α helices. The predicted IpaA-VBSs were arranged around a helical wheel, which revealed amphipathic helices having a hydrophilic basic face, and a hydrophobic face. Hydrophobic residues are boxed. (E) Either IpaA-VBS or -VBS2 is sufficient to bind to vinculin. Media from cultures of the indicated strains of S. flexneri were harvested and analyzed for the expression of S. flexneri's invasin proteins, which were detected by staining SDS-PAGE gels with Coomassie blue (top). Samples were then blotted and levels of wild-type and mutant IpaA proteins were determined by Western blotting with IpaA-specific antibody (middle). Samples were also blotted with I125-labeled vinculin using a vinculin overlay assay. Note that vinculin could bind to IpaA, IpaA-ΔVBS, and -ΔVBS2, but not to IpaA-ΔCterm, which lacks both of the VBSs of IpaA. The mxiD deletion (type III secretion system defective) does not secrete any of the invasin proteins (Perdomo et al., 1994).