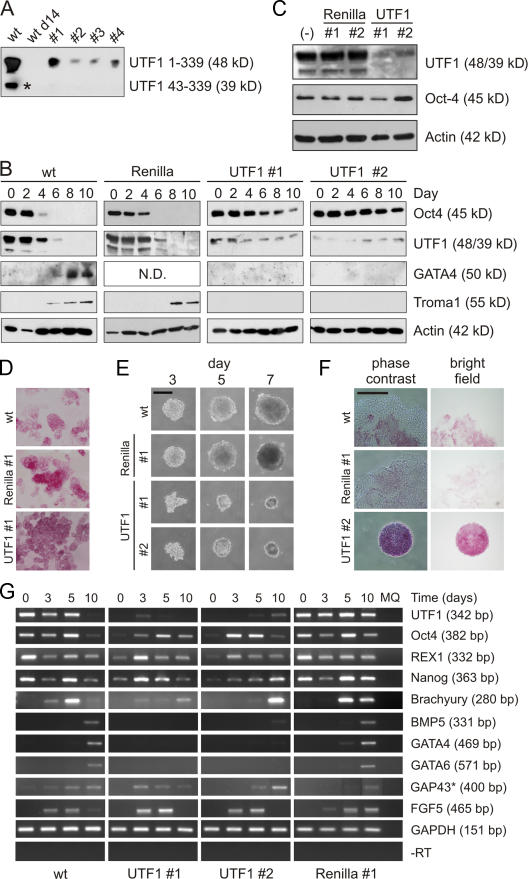
Figure 1.
UTF1 is involved in the differentiation of EC and ES cells. (A) UTF1 expression in P19CL6 EC cells (wt), 14-d DMSO-differentiated EC cells (wt d14), and four independent UTF1 EC KD clones (UTF1 #1–#4). The asterisk indicates a shorter variant of mUTF1 (aa 43–339) generated by transcription from an alternative start site (Nishimoto et al., 2001). (B) DMSO-induced differentiation of wt, Renilla luciferase KD (Renilla), and UTF1 KD (UTF1 #1 and #2) EC cells. Cell lysates were analyzed with antibodies against Oct4, UTF1, GATA4, and Troma1. Actin staining was performed as a loading control. (C) Western analysis of wt, Renilla luciferase KD (Renilla #1 and #2), and UTF1 KD (UTF1 #1 and #2) IB10 ES cells. Cell lysates were analyzed with antibodies against UTF1 and Oct4. Actin levels were determined to correct for gel loading. (D) Brightfield images of wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #1) ES cells stained for AP activity. (E) Phase-contrast images of EBs from wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #1 and #2) ES cells after 3, 5, and 7 d. (F) Phase-contrast and brightfield images of day 8 EBs from wt, Renilla luciferase KD (Renilla #1), and UTF1 KD (UTF1 #2) ES cells stained for AP activity. (G) Expression levels of markers for ES cells (UTF1, Oct4, REX1, and Nanog), ectoderm (FGF5 and GAP43), mesoderm (Brachyury and BMP5), and endoderm (GATA4 and GATA6) were measured by semiquantitative RT-PCR in undifferentiated ES cells and EBs cultured for 3, 5, and 10 d. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as a control. In the −RT lanes, reverse transcriptase was omitted from the reverse transcriptase reactions to control for genomic DNA contamination and amplified using glyceraldehyde-3-phosphate dehydrogenase primers. A representative experiment is depicted. The asterisk indicates that the 5- and 10-d GAP43 RT-PCR products were not loaded in adjacent lanes. Bars, 250 μm.