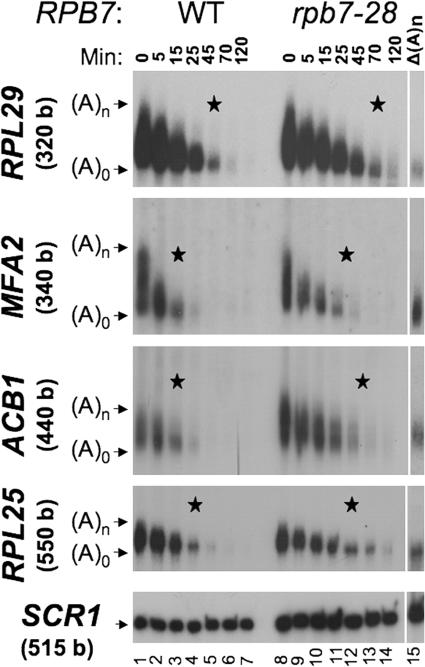
Figure 3.
Rpb7p is required for efficient deadenylation and subsequent decay of both PBF and non-PBF mRNA. Cells were shifted rapidly to 42°C to block transcription and to inactivate the ts Rpb7-28p. RNA was extracted at the indicated time points post-shift and analyzed by the polyacrylamide Northern technique (Sachs and Davis, 1989). The same membrane was hybridized sequentially with the indicated probes. Actual lengths (relative to size marker) of the indicated mRNAs (without the poly(A) tail) are indicated in parentheses next to the gene name on the left. Lane “Δ(A)n” shows the position of fully deadenylated RNA. This RNA was obtained by hybridizing RNA sample from time point 0 with oligo(dT), followed by digestion of the poly(A) tail by RNase H. The star indicates the time point when deadenylation seems to be complete. We suspect that in some cases complete deadenylation occurred in between two time points. In these particular cases, the star is placed between lanes. SCR1 RNA (a Pol III transcript) serves to demonstrate equal loading.