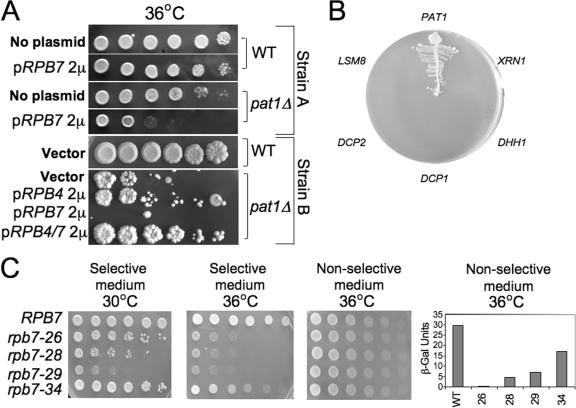
Figure 6.
Rpb7p interacts with Pat1p; the extent of interaction correlates with the capacity of Rpb7p to stimulate mRNA decay. (A) High copy effect of the indicated genes on the proliferation capacity of pat1Δ cells or their WT counterparts. Two strain backgrounds were examined. WT strain (yMC229) and its isogenic pat1Δ strain (yMC363) are designated “Strain A”; WT (yMC269) or its pat1Δ derivative yMC272 are designated “Strain B”. Cells carrying high copy plasmid expressing the indicated genes were spotted in fivefold serial dilutions onto selective plates. Cells were allowed to grow at the nonpermissive temperature for pat1Δ cells, 36°C. (B) Two-hybrid interactions between Rpb7p as the bait and genes whose products are involved in mRNA decay as prey were performed as described in Materials and methods. Only 6 of 20 genes tested are shown as indicated around the plate. The other genes that did not exhibit interactions with the Rpb7p-DBD and are not shown here are Lsm1p, Lsm3p–Lsm7p, Ccr4p, Not1p, Caf1p, Pan2p, Pan3p, Pop2p, Edc1p, Edc2p, Edc3p, and Pab1p. Lsm2p showed weak interaction (not depicted). Equal amount of cells, carrying the indicated prey plasmids, were streaked onto an indicator plate as described in Materials and methods. We then verified that the growth on the indicator plates was dependent on both plasmids by evicting one plasmid at a time from the positive clones (not depicted). (C) Two-hybrid interaction between various mutant forms of Rpb7p-DBD and Pat1p-AD. Cells were spotted in threefold serial dilutions, starting with 106 cells/spot, on an indicator plate as in B (selective conditions). Plates were incubated for 3 d at the indicated temperatures. To demonstrate spotting of equal amounts of cells, cells were spotted on a nonselective plate that allowed growth of cells that carry both the bait and prey plasmids independently of the two-hybrid interactions (using medium lacking only leucine and tryptophane). β-galactosidase (β-Gal) values, quantitative means to determine interactions (Uetz et al., 2000), are indicated on the right. They reflect the average values determined from two independent experiments (variations were <15%). Background values obtained with cells expressing only the bait plasmid were subtracted. Levels of the various mutant forms of Rpb7p-DBD were found to be comparable to that of the WT Rpb7-DBD (not depicted).