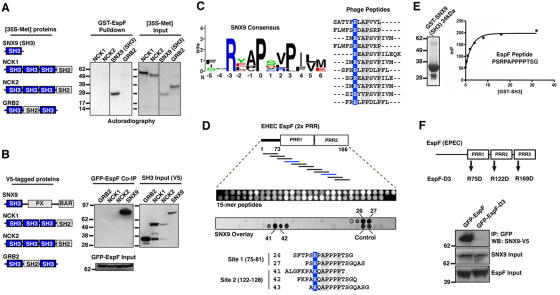
Figure 2.
Identification of motifs required for direct EspF and SNX9 interactions. (A) Glutathione-Sepharose pull-down with 10 μg of GST-EspF (residues 48–206) mixed with the [35S]-methionine proteins indicated (left diagram). Autoradiograph of GST pulldown (left) and of 1/20th input of 35S-labeled NCK1, NCK2, SNX9 (residues 1–111), and GRB2 is shown (right). (B) HEK293A cells were cotransfected with EGFP-EspF and V5-tagged proteins indicated (left diagram). Anti-GFP immunoprecipitations (IP) were probed by V5 immunoblot (IB) (left). Cell lysates were probed by V5 or GFP immunoblot to show input levels. (C) Logos plot of the SNX9 binding consensus sequence derived by phage display experiments (left). Alignment of 13 unique SNX9 binding sequences used to derive the consensus is shown. The invariant arginine (blue) and highly conserved residues (gray) are highlighted. (D) Peptide array analysis of the SNX9-binding sites on EspF. Top: diagram of EspF residues 1–166 used for the peptide scanning experiments. Middle: ultraviolet (UV) illumination shows the qualitative amount of each peptide synthesized (top). Bottom: solid-phase binding of 35S-SNX9 to 15-mer EspF peptides was assessed by autoradiography. An alignment of EspF-binding peptides from two SNX9 binding series is shown. (E) Saturation binding curves were generated with increasing concentrations of GST-SNX9-SH3 (left) to a fixed concentration of EspF peptide by fluorescence polarization (see Materials and methods). (F) HEK293A cells were cotransfected with EGFP-EspF or triple mutant EGFP-EspF-D3 (top diagram) and V5-tagged SNX9. Anti-GFP immunoprecipitations (IP) were probed by V5 immunoblot (IB) (top panel). Cell lysates were probed by V5 or GFP immunoblot to show input levels (bottom two panels).