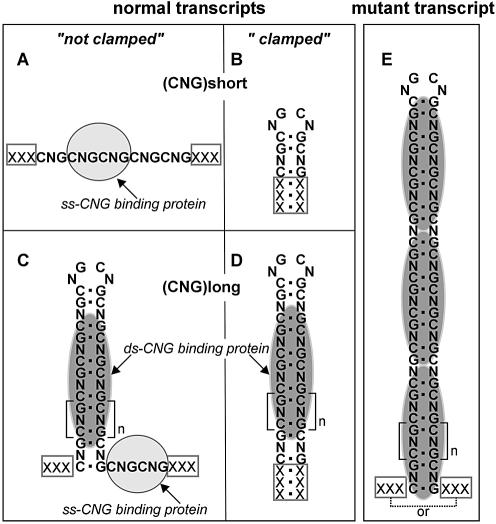
Figure 9.
A model depicting the structures and protein binding properties of the CNG repeats in transcripts. Different types of structures formed by the repeats and their flanking sequences in transcripts as well as two different types of the CNG repeat binding proteins are schematically shown. The ability of the CNG repeats to form hairpin structures depends on two factors: the presence of a stable duplex structure (a ‘clamp’) formed by the natural sequences flanking the repeats in their host transcripts, and the repeat length. (A) Short non-clamped repeats do not form hairpin structures and bind the ssCNG repeat binding protein (gray oval). (B) Short clamped repeats form the hairpin structure which may not have a protein binding capacity. (C) Long normal (>20) non-clamped repeats form stable ‘slippery’ hairpin structures that may bind both the dsCNG repeat binding protein (dark gray oval) to the hairpin stem, and ssCNG repeat binding protein to the protruding repeat tail as demonstrated for the CUG repeat transcript and the CUG-BP (8). (D) Long normal clamped repeats form stable hairpin structures that bind the dsCNG repeat binding protein only. (E) An expanded repeat has the structure and protein binding specificity similar to that of the long normal repeats (C) or (D) but binds the dsCNG repeat binding protein in a length-dependent manner.