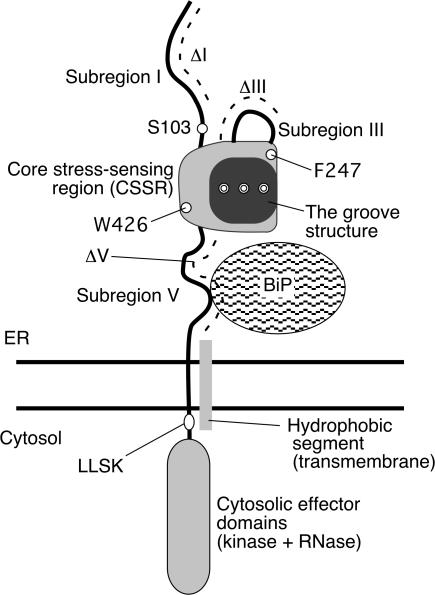
Figure 1.
Structure of yeast Ire1 and mutations used in this study. Structure of the luminal domain according to Kimata et al. (2004), Oikawa et al. (2005), and Credle et al. (2005) is illustrated. Subregions I (aa 32–111), III (aa 243–272), and V (aa 455–524) are loosely folded, whereas subregions II (aa 112–242) and IV (aa 273–454) form the tightly folded CSSR. The position of a hydrophobic segment (aa 527–570) that is deduced to be the transmembrane domain is also shown. The dashed lines indicate positions of amino acid residues deleted in ΔI (aa 32–91), ΔIII (aa 253–272), and ΔV (aa 463–524) mutants, respectively. The positions of another deletion mutation, 567LLSK570, and point mutations S103, F247, and W246 are also indicated. Double circles respectively represent M229, F285, and Y301, which are simultaneously replaced by Ala in Figs. 5 E and 7. Because we now assign the initiation methionine according to the data from the Saccharomyces genome database (http://www.yeastgenome.org/), the amino acid numbers of Ire1 in the present paper differ by 7 aa from those in two of our previous papers (Kimata et al., 2004; Oikawa et al., 2005).