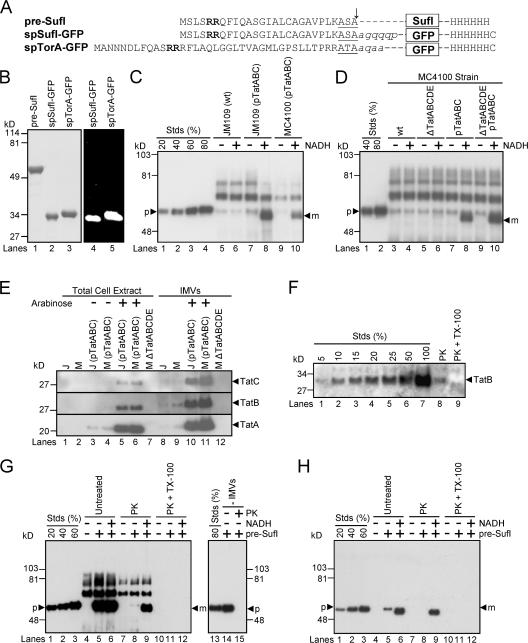
Figure 1.
Precursor proteins, transport efficiencies, TatABC expression levels, and membrane orientation in in vitro Tat transport assays. (A) The precursor proteins used in the transport assays. The twin-arginine motifs (bold), signal peptidase recognition sequences (underlined) and signal sequence cleavage sites (arrow) are identified. Lowercase, italicized letters are residues of the mature SufI and TorA domains that have been retained in the GFP fusions. (B) Purity and fluorescence of the precursor proteins. Purified precursor proteins were visualized in SDS-PAGE gels by Coomassie blue staining (left) and the fluorescence emission upon UV excitation (right). (C) A standard in vitro transport assay. In this anti-SufI immunoblot, known amounts of pre-SufI (lanes 1–4), marked as a percentage of the precursor protein (p) added to the transport reactions, allowed quantification of the transport efficiency. Samples were proteinase K treated after 30 min at 37°C. NADH (4 mM) promoted the appearance of protease-protected mature-length protein (m). Higher molecular weight bands result from immuno-crossreactivity with unknown endogenous proteins (see G). Under some conditions, minute quantities of precursor were detectable in the absence of NADH; control experiments indicate that this was likely due to incomplete protease digestion, and this amount was always subtracted from that observed in the presence of NADH for accurate quantification of transport. Efficient transport required overexpression of TatABC, and was typically higher in JM109(pTatABC) than in MC4100(pTatABC), here quantified as 73 and 45%, respectively. Lane 8 was considered standard assay conditions. [pre-SufI] = 50 nM; [IMV] = 5 (A 280). (D) Transport efficiencies of different MC4100-derived strains. This anti-SufI immunoblot demonstrates that overexpression of TatABC promoted efficient protein transport (compare lanes 4 and 8), and that TatD and TatE were not required for efficient transport (compare lanes 6 and 10). Same conditions as panel C. (E) TatABC overexpression. Expression of TatA, TatB, and TatC proteins was induced from the pTatABC plasmid in JM109 (J) and MC4100 (M) strains with 0.7% arabinose, as indicated. All three proteins were retained in IMVs. Five microliters of total cell extracts (A 600 = 4.4) were loaded per lane; 1 μl of IMVs (A 280 = 50) was loaded per lane. (F) TatB accessibility assay. The percentage of inside-out oriented vesicles in IMV preparations was determined by the protease accessibility of the TatB cytoplasmic domain. This anti-TatB immunoblot shows different amounts of control IMVs for quantification purposes (lanes 1–7), where the 100% lane represents the amount of TatB present before proteinase K (PK) treatment. After protease treatment (lane 8), ∼10% of the original TatB remained, and upon protease treatment in the presence of 0.05% TX-100 (lane 9), all the TatB was digested. These data indicate that the vesicles in this IMV preparation were ∼90% inside-out. (G) Protease digestion of mature SufI after membrane permeabilization. The left anti-SufI immunoblot shows that addition of 0.05% Triton X-100 (TX-100) during proteinase K (PK) treatment under standard assay conditions (see panel C) resulted in complete loss of the mature SufI protein band (compare lanes 9 and 12). The right anti-SufI immunoblot shows that PK addition resulted in complete digestion of pre-SufI. (H) Same experiment as panel G, but using 6xHis antibodies. Due to the lower detection efficiency of the 6xHis antibodies, samples were sedimented and gel loads (based on IMV amount) were 16 times higher than in panel G.